Statistical analysis and plots
From: Comparative transcriptomics of social insect queen pheromones
2019-03-07
Last updated: 2019-03-07
Checks: 6 0
Knit directory: queen-pheromone-RNAseq/
This reproducible R Markdown analysis was created with workflowr (version 1.2.0). The Report tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190307) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: bioinformatics_scripts/data/
Ignored: bioinformatics_scripts/ref/
Ignored: data/.DS_Store
Ignored: data/apis_gene_comparisons/.DS_Store
Ignored: data/apis_gene_comparisons/wojciechowski_histone_data/.DS_Store
Ignored: data/apis_gene_comparisons/wojciechowski_histone_data/GSE110640_RAW/.DS_Store
Ignored: data/apis_gene_comparisons/wojciechowski_histone_data/GSE110641_RAW/.DS_Store
Ignored: data/component spreadsheets of queen_pheromone.db/.DS_Store
Ignored: docs/figure/pdf_supplementary_material.Rmd/
Ignored: figures/
Ignored: manuscript/
Ignored: supplement/
Unstaged changes:
Deleted: bioinformatics_scripts/data
Deleted: bioinformatics_scripts/ref
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 7d083c0 | lukeholman | 2019-03-07 | new theme |
| html | d5a6a73 | lukeholman | 2019-03-07 | Build site. |
| html | f260a62 | Luke Holman | 2019-03-07 | Build site. |
| Rmd | 934cda8 | Luke Holman | 2019-03-07 | First commit of new website structure |
This document was written in R Markdown, and translated into html using the R package knitr. Press the buttons labelled Code to show or hide the R code used to produce each table, plot or statistical result. You can also select Show all code at the top of the page.
Load R libraries (install first from CRAN or Bioconductor)
library(WGCNA) # Gene networks - needs 'impute' dependency: source("https://bioconductor.org/biocLite.R"); biocLite("impute")
library(RSQLite) # Access SQLite databases
library(reshape2) # data tidying (melt)
library(dplyr) # data tidying
library(tidyr) # data tidying
library(purrr) # manipulate lists
library(stringr) # string manipulation
library(ggplot2) # for plots
library(ggrepel) # for plots
library(ggdendro) # for plots
library(gridExtra) # for plots
library(grid) # for plots
library(RColorBrewer) # for plots
library(ggjoy) # for plots
library(gplots) # Venn diagram
library(ecodist) # for nmds
library(MuMIn) # for model comparison
library(sva) # for ComBat function; install via source("https://bioconductor.org/biocLite.R"); biocLite("sva")
library(pander) # for nice tables
library(kableExtra) # For scrollable tables
library(clusterProfiler) # for enrichment tests; source("https://bioconductor.org/biocLite.R"); biocLite("clusterProfiler")
library(fgsea) # for enrichment tests; source("https://bioconductor.org/biocLite.R"); biocLite("fgsea")
select <- dplyr::select
filter <- dplyr::filter
rename <- dplyr::rename
kable.table <- function(df) {
kable(df, "html") %>%
kable_styling() %>%
scroll_box(height = "300px")
}
# Open database connections (Sasha uses SQL, Luke prefers dplyr)
db <- dbConnect(SQLite(), dbname="data/queen_pheromone.db")
my_db <- src_sqlite("data/queen_pheromone.db")
# These 4 samples should NOT be used (See below). They were also removed in all non-R analyses (e.g. differential gene expression analyses using EBseq)
bad.samples <- c("lf1", "ln1", "ln12", "lf12")First let’s check for and remove strongly outlying samples
# Define a function to get gene expression data for a given set of orthologous genes. We define orthologous genes as those that are each other's reciprocal best BLAST. The bad.samples argument can be used to remove some named samples. By default this function logs the expression data (using log10). It returns a list with two elements: the first element is a matrix of expression data (rows = samples, cols = genes), and the second is a data frame giving the species-specific names of the orthologous genes
make.OGGs <- function(species, bad.samples = NULL, log.data = T){
# set up forward mappings, e.g. "am2bt", "am2lf", "am2ln"
forward.mappings <- paste(species[1], "2", species[2:length(species)], sep = "")
# and reverse mappings, e.g. "bt2am", "lf2am", "ln2am"
backward.mappings <- paste(species[2:length(species)], "2", species[1], sep = "")
items <- list() # declare empty list
for(i in 1:length(forward.mappings)){
# make a table with 3 columns: first column has species 1 gene,
# second column has species 2 gene in forward mapping,
# third column has species 2 gene in reverse mapping (this can be NA, or different to col 2)
# we want the rows where cols 2 and 3 are the same, indicating reciprocity in the BLAST
focal <- left_join(
tbl(my_db, forward.mappings[i]) %>%
dplyr::select(-evalue), # get the two mappings and
tbl(my_db, backward.mappings[i]) %>%
dplyr::select(-evalue), # merge by species 1 column
by = species[1]
) %>%
collect(n=Inf) %>% as.data.frame # collect it all and convert to df
# Get the RBB rows, and keep the two relevant columns
focal <- focal[!is.na(focal[,3]), ]
items[[i]] <- focal[focal[,2] == focal[,3], 1:2]
}
rbbs <- items[[1]] # If 3 or 4 species, successively merge the results
if(length(items) > 1) rbbs <- left_join(rbbs, items[[2]], by = species[1])
if(length(items) > 2) rbbs <- left_join(rbbs, items[[3]], by = species[1])
# Throw out species1 genes that do not have a RBB in all species
rbbs <- rbbs[complete.cases(rbbs), ]
names(rbbs) <- gsub("[.]x", "", names(rbbs)) # tidy the row and column names
rownames(rbbs) <- NULL
# Make sure the columns are ordered as in 'species'
rbbs <- rbbs[, match(names(rbbs), species)]
# We know have a list of the names of all the ortholgous genes in each species
# Now we use these names to look up the gene expression data for each ortholog
expression.tables <- paste("rsem_", species, sep = "")
for(i in 1:length(species)){
focal.expression <- tbl(my_db, expression.tables[i]) %>%
collect(n=Inf) %>% as.data.frame()
names(focal.expression)[names(focal.expression) == "gene"] <- species[i]
rbbs <- left_join(rbbs, focal.expression, by = species[i])
}
gene.name.mappings <- rbbs[, names(rbbs) %in% species] # save gene name mappings in separate DF
rownames(rbbs) <- rbbs[,1] # Use the gene names for species 1 as row names
rbbs <- rbbs[, !(names(rbbs) %in% species)] # remove gene name columns
rbbs <- t(as.matrix(rbbs))
if(log.data) rbbs <- log10(1 + rbbs)
if(!is.null(bad.samples)) rbbs <- rbbs[!(rownames(rbbs) %in% bad.samples), ]
# Discard genes where NAs appear
gene.name.mappings <- gene.name.mappings[!(is.na(colSums(rbbs))), ]
rbbs <- rbbs[, !(is.na(colSums(rbbs)))]
# discard genes where expression is zero for all samples in 1 or more species
spp <- str_replace_all(rownames(rbbs), "[:digit:]", "")
to.keep <- rep(TRUE, ncol(rbbs))
for(i in 1:ncol(rbbs)){
if(min(as.numeric(tapply(rbbs[,i], spp, sum))) == 0) to.keep[i] <- FALSE
}
rbbs <- rbbs[, to.keep]
gene.name.mappings <- gene.name.mappings[to.keep, ]
list(tpm = rbbs, gene.mappings = gene.name.mappings)
}4 of the Lasius samples are highly different to all of the rest
Closer inspection reveals that they have zeros for many of the transcripts, so perhaps they had low abundance libraries.
set.seed(1) # nmds involves random numbers, so make this plot reproducible
expression.data <- make.OGGs(c("am", "bt", "ln", "lf"))[[1]]
treatments <- tbl(my_db, "treatments") %>% as.data.frame()
shhh <- capture.output(nmds.output <- dist(expression.data) %>% nmds())
fig_S1 <- data.frame(id = rownames(expression.data), nmds.output$conf[[length(nmds.output$conf)]], stringsAsFactors = F) %>%
left_join(treatments, by = "id") %>%
rename(Species = species, Treatment = treatment) %>%
ggplot(aes(X1,X2, shape = Species)) +
geom_point(aes(colour = Treatment)) +
geom_text_repel(aes(label = id), size=3.6) +
xlab("NMDS 1") + ylab("NMDS 2")
saveRDS(fig_S1, file = "supplement/fig_S1.rds")
fig_S1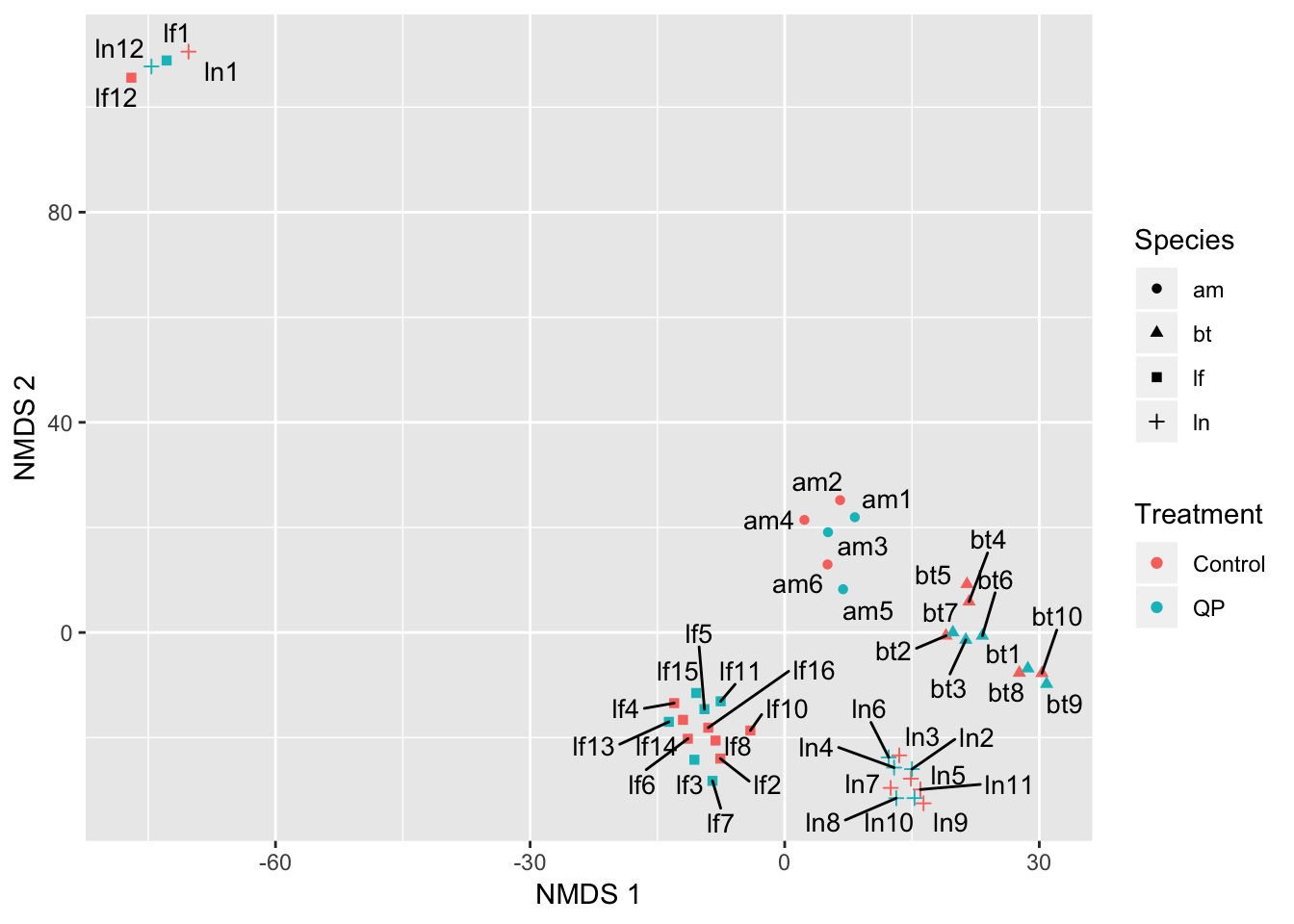
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Figure S1: After reducing the transcriptome data to two axes using non-metric multidimensional scaling, four Lasius samples were clear outliers.
set.seed(1) # nmds involves random numbers, so make this plot reproducible
expression.data <- make.OGGs(c("am", "bt", "ln", "lf"), bad.samples = bad.samples)[[1]]
shhh <- capture.output(nmds.output <- dist(expression.data) %>% nmds())
fig_S2 <- data.frame(id = rownames(expression.data),
nmds.output$conf[[length(nmds.output$conf)]],
stringsAsFactors = F) %>%
left_join(treatments, by = "id") %>%
rename(Species = species, Treatment = treatment) %>%
ggplot(aes(X1,X2)) +
geom_point(aes(shape = Species, colour = Treatment)) +
geom_text_repel(aes(label = id), size=3.6) +
xlab("NMDS 1") + ylab("NMDS 2")
saveRDS(fig_S2, file = "supplement/fig_S2.rds")
fig_S2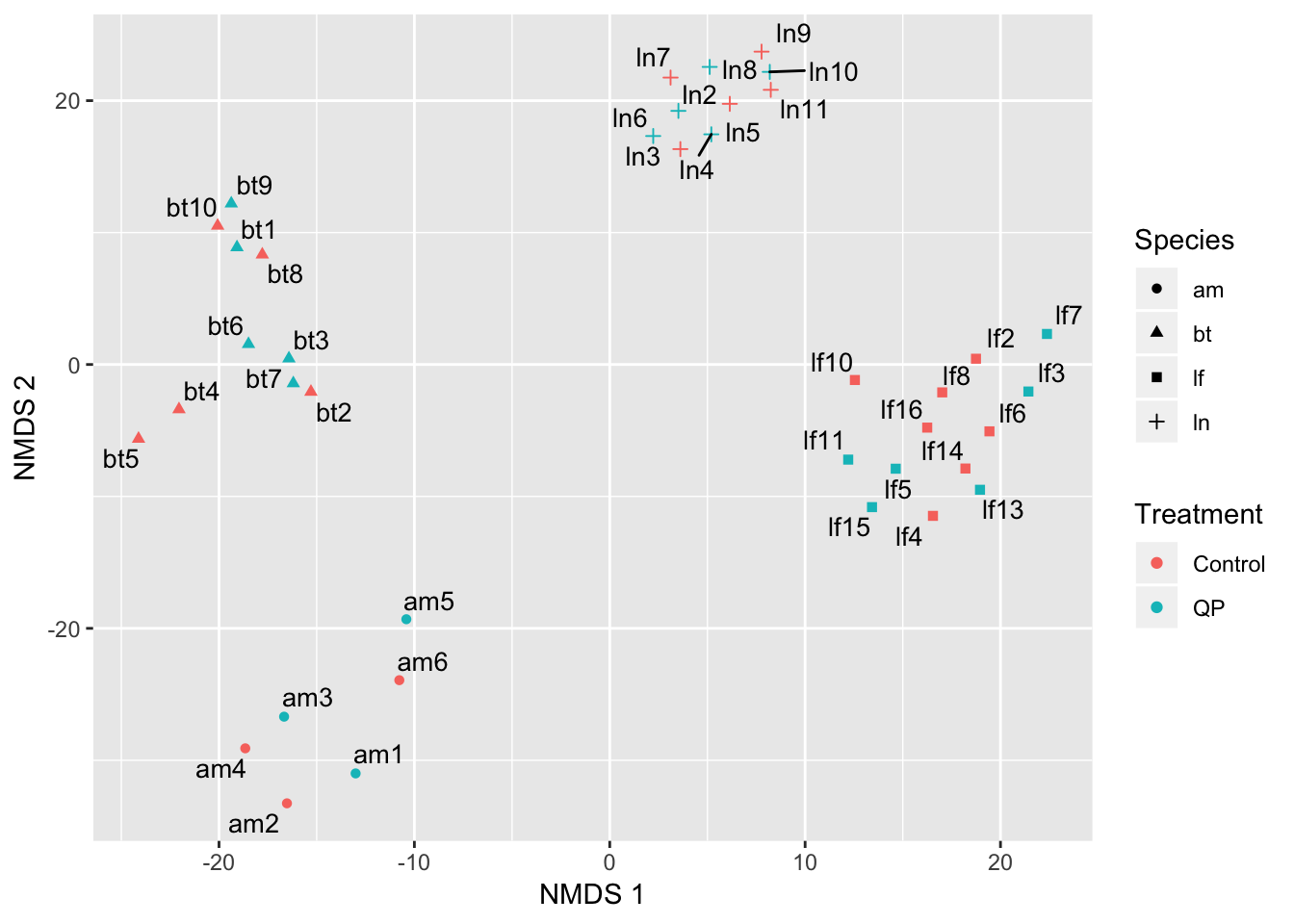
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Figure S2: With the four problematic samples removed, the samples cluster according to species with no obvious outliers.
Table of sample sizes
Table S1: Number of sequencing libraries for each combination of species and treatment, after removing the four problematic libraries. Each library was prepared from a pool containing equal amounts of cDNA from five individual workers, taken from the same colony.
sample.size.table <- treatments[treatments$id %in% rownames(expression.data),] %>%
group_by(species, treatment) %>%
summarise(n = n()) %>% as.data.frame()
names(sample.size.table) <- c("Species", "Treatment", "Number of RNAseq libraries")
saveRDS(sample.size.table, file = "supplement/tab_S1.rds")
sample.size.table %>% pander()| Species | Treatment | Number of RNAseq libraries |
|---|---|---|
| am | Control | 3 |
| am | QP | 3 |
| bt | Control | 5 |
| bt | QP | 5 |
| lf | Control | 7 |
| lf | QP | 6 |
| ln | Control | 5 |
| ln | QP | 5 |
Lists of statistically significant differentially expressed genes
Click the tabs to see the gene lists for each of the four species.
apis.de <- suppressMessages(tbl(my_db, "ebseq_padj_gene_am") %>%
dplyr::select(gene, PostFC) %>%
left_join(tbl(my_db, "bee_names")) %>%
collect() %>%
mutate(PostFC = round(log2(PostFC), 3)) %>%
dplyr::select(gene, name, PostFC) %>%
rename(Gene=gene, Name=name, Log2_FC = PostFC) %>%
arrange(-abs(Log2_FC))) %>% as.data.frame()
bombus.de <- suppressMessages(tbl(my_db, "ebseq_padj_gene_bt") %>%
dplyr::select(gene, PostFC) %>%
left_join(tbl(my_db, "bt2am") %>% rename(gene=bt)) %>%
left_join(tbl(my_db, "bee_names") %>% rename(am=gene)) %>%
collect() %>%
mutate(PostFC = round(log2(PostFC), 3), name = replace(name, is.na(name), " "),
am = replace(am, is.na(am), " ")) %>%
dplyr::select(gene, am, name, PostFC) %>%
rename(Gene=gene, Apis_BLAST=am, Name=name, Log2_FC = PostFC) %>%
arrange(-abs(Log2_FC))) %>% as.data.frame()
flavus.de <- suppressMessages(tbl(my_db, "ebseq_padj_gene_lf") %>%
dplyr::select(gene, PostFC) %>%
left_join(tbl(my_db, "lf2am") %>% rename(gene=lf)) %>%
left_join(tbl(my_db, "bee_names") %>% rename(am=gene)) %>%
collect() %>%
mutate(PostFC = round(log2(PostFC), 3), name = replace(name, is.na(name), " "),
am = replace(am, is.na(am), " ")) %>%
dplyr::select(gene, am, name, PostFC) %>%
rename(Gene=gene, Apis_BLAST=am, Name=name, Log2_FC = PostFC) %>%
arrange(-abs(Log2_FC))) %>% as.data.frame()
niger.de <- suppressMessages(tbl(my_db, "ebseq_padj_gene_ln") %>%
dplyr::select(gene, PostFC) %>%
left_join(tbl(my_db, "ln2am") %>% rename(gene=ln)) %>%
left_join(tbl(my_db, "bee_names") %>% rename(am=gene)) %>%
collect() %>%
mutate(PostFC = round(log2(PostFC), 3), name = replace(name, is.na(name), " "),
am = replace(am, is.na(am), " ")) %>%
dplyr::select(gene, am, name, PostFC) %>%
rename(Gene=gene, Apis_BLAST=am, Name=name, Log2_FC = PostFC) %>%
arrange(-abs(Log2_FC))) %>% as.data.frame()
names(apis.de) <- gsub("_", " ", names(apis.de))
names(bombus.de) <- gsub("_", " ", names(bombus.de))
names(flavus.de) <- gsub("_", " ", names(flavus.de))
names(niger.de) <- gsub("_", " ", names(niger.de))Apis mellifera
Table S2: List of the 322 significantly differentially expressed genes (EBseq; FDR-corrected posterior probability of differential expression p < 0.05) in Apis mellifera, listed in order of fold change in gene expression on a Log\(_2\) scale. Positive fold change values indicate higher expression in the control, while negative values indicate higher expression in the queen pheromone treatment.
saveRDS(apis.de, file = "supplement/tab_S2.rds")
kable.table(apis.de)| Gene | Name | Log2 FC |
|---|---|---|
| GB55204 | Major royal jelly protein 3 | 6.480 |
| GB51373 | bypass of stop codon protein 1-like | 5.385 |
| GB50604 | uncharacterized protein LOC724113 | 3.775 |
| 102655911 | uncharacterized protein LOC102655911 | -3.376 |
| GB49819 | branched-chain-amino-acid aminotransferase, cytosolic-like | 2.657 |
| 102656917 | uncharacterized LOC102656917, transcript variant X1 | 2.609 |
| GB54417 | dehydrogenase/reductase SDR family member 11-like isoform X1 | 2.242 |
| GB45565 | chymotrypsin-2 | 2.052 |
| GB53886 | protein G12-like isoform X4 | 1.983 |
| 100576536 | uncharacterized protein LOC100576536 | 1.945 |
| GB41540 | venom carboxylesterase-6-like | -1.721 |
| GB54690 | uncharacterized protein LOC408547 | -1.583 |
| GB54150 | uncharacterized protein LOC408462 | -1.560 |
| GB43639 | uncharacterized protein LOC100577506 isoform X1 | -1.557 |
| GB49548 | serine/threonine-protein phosphatase 2B catalytic subunit 3-like isoform X11 | -1.455 |
| GB49878 | probable cytochrome P450 6a14 isoformX1 | 1.393 |
| GB53414 | serine/threonine-protein kinase ICK-like isoform X2 | 1.357 |
| 102654781 | protein G12-like | 1.324 |
| 102656058 | uncharacterized protein PF11_0213-like | -1.311 |
| GB53957 | U6 snRNA-associated Sm-like protein LSm1-like | 1.276 |
| GB50413 | protein TBRG4-like isoform X1 | -1.261 |
| 102653931 | uncharacterized LOC102653931, transcript variant X2 | -1.258 |
| GB53876 | interaptin-like | -1.209 |
| GB42705 | protein archease-like | 1.184 |
| 101664701 | PI-PLC X domain-containing protein 1-like isoform X1 | 1.143 |
| GB42523 | uncharacterized LOC100577781, transcript variant X2 | -1.130 |
| 102654405 | protein G12-like | 1.115 |
| 100578075 | uncharacterized LOC100578075 | -1.110 |
| GB52251 | multifunctional protein ADE2, transcript variant X2 | 1.088 |
| GB40764 | uncharacterized protein LOC414021 isoform X7 | -1.086 |
| GB55648 | Down syndrome cell adhesion molecule-like protein Dscam2-like isoform X7 | -1.084 |
| 726446 | uncharacterized protein LOC726446 | -1.063 |
| GB54467 | probable G-protein coupled receptor 52 isoform 1 | -1.060 |
| GB46985 | 60S ribosomal protein L12 isoform X1 | 1.045 |
| GB55191 | uncharacterized protein LOC100576289 | -1.045 |
| GB54890 | kynurenine 3-monooxygenase isoform X2 | 1.016 |
| GB55640 | retinol dehydrogenase 12-like | -1.006 |
| 724802 | protein Asterix-like | 0.996 |
| GB40010 | titin-like isoform X2 | -0.967 |
| 102654949 | uncharacterized protein LOC102654949 | 0.967 |
| GB43234 | histone deacetylase 5 isoform X8 | -0.965 |
| GB52266 | furin-like protease 2-like | -0.927 |
| GB41706 | ice-structuring glycoprotein-like | -0.926 |
| GB42673 | retinol dehydrogenase 10-A-like isoform X4 | 0.923 |
| GB55030 | uncharacterized protein LOC725074 | 0.921 |
| 551123 | RNA-binding protein Musashi homolog Rbp6-like isoform X1 | -0.920 |
| 409728 | 40S ribosomal protein S5 isoform X1 | 0.917 |
| GB45028 | venom dipeptidyl peptidase 4 | -0.898 |
| GB53422 | ufm1-specific protease 1-like isoform X2 | 0.892 |
| GB48933 | methenyltetrahydrofolate synthase domain-containing protein-like | -0.876 |
| GB53077 | cysteine-rich protein 1-like | 0.871 |
| GB41301 | annexin-B9-like | 0.862 |
| GB51748 | dentin sialophosphoprotein | -0.861 |
| 102654594 | WD repeat-containing protein 18-like | 0.852 |
| GB50356 | 60S acidic ribosomal protein P2 | 0.837 |
| GB44091 | LOW QUALITY PROTEIN: uncharacterized protein LOC408779 | -0.836 |
| GB41151 | protein MNN4-like | -0.834 |
| 409202 | ribosomal protein S9, transcript variant X2 | 0.834 |
| GB44340 | small ubiquitin-related modifier 3 isoform 1 | 0.833 |
| GB49173 | 4-aminobutyrate aminotransferase, mitochondrial-like isoform X2 | -0.826 |
| GB40769 | dehydrogenase/reductase SDR family member 11-like | 0.823 |
| GB54243 | LOW QUALITY PROTEIN: carbonyl reductase [NADPH] 1-like | 0.819 |
| 724531 | 40S ribosomal protein S28-like | 0.812 |
| GB51744 | uncharacterized protein LOC724439 | 0.811 |
| GB51947 | uncharacterized protein LOC724835 isoform X2 | -0.807 |
| GB40875 | 60S ribosomal protein L10 isoform X1 | 0.806 |
| 102655694 | glutathione S-transferase-like | 0.803 |
| GB55827 | 40S ribosomal protein S21-like isoform X1 | 0.801 |
| 102654426 | 60S ribosomal protein L18-like | 0.801 |
| GB49988 | SRR1-like protein-like isoform X2 | -0.800 |
| GB55963 | uncharacterized protein LOC725224 isoform X1 | -0.797 |
| GB50867 | cell differentiation protein RCD1 homolog isoform X2 | 0.794 |
| GB43256 | ATP-binding cassette sub-family D member 1-like | 0.792 |
| GB41211 | ATP-binding cassette sub-family E member 1 | 0.787 |
| GB52314 | gamma-tubulin complex component 4 | -0.785 |
| 102654251 | uncharacterized protein LOC102654251 | -0.782 |
| 726860 | cytochrome b5-like isoform 1 | 0.778 |
| GB46039 | tubulin alpha-1 chain-like | 0.773 |
| GB53358 | protein transport protein Sec61 subunit gamma-like isoform X3 | 0.768 |
| GB48699 | 60S ribosomal protein L11-like | 0.767 |
| GB44311 | actin related protein 1 | 0.762 |
| 102655603 | transmembrane emp24 domain-containing protein 7-like | 0.762 |
| GB49170 | 40S ribosomal protein S15Aa-like isoform 1 | 0.758 |
| GB47736 | alkyldihydroxyacetonephosphate synthase-like | 0.753 |
| GB49013 | RNA-binding protein 8A | 0.752 |
| 724485 | probable small nuclear ribonucleoprotein E-like | 0.749 |
| GB53000 | ubiquitin-60S ribosomal protein L40 isoform 2 | 0.749 |
| 725936 | titin-like | -0.748 |
| GB50158 | 60S ribosomal protein L4 isoform 1 | 0.748 |
| GB52432 | KN motif and ankyrin repeat domain-containing protein 3-like isoform X3 | -0.746 |
| 724757 | histone H4-like | 0.745 |
| GB53219 | 40S ribosomal protein S17 | 0.742 |
| 100577623 | putative uncharacterized protein DDB_G0282133-like isoform X2 | -0.742 |
| GB51038 | 60S ribosomal protein L23 | 0.739 |
| GB40284 | cytochrome P450 6a2 | 0.737 |
| GB50709 | 40S ribosomal protein S19a | 0.735 |
| GB50977 | probable tubulin polyglutamylase TTLL2-like | -0.734 |
| GB42537 | 40S ribosomal protein S15 | 0.734 |
| GB42467 | phospholipase B1, membrane-associated-like isoform X2 | 0.733 |
| GB41886 | protein transport protein Sec61 subunit alpha isoform 2 | 0.729 |
| GB51201 | 40S ribosomal protein S12 isoform X1 | 0.724 |
| GB55183 | ankyrin repeat domain-containing protein SOWAHB-like isoform X5 | -0.724 |
| GB54814 | 60S ribosomal protein L31 isoform 1 | 0.721 |
| GB49159 | probable nuclear transport factor 2-like isoform 3 | 0.717 |
| GB52512 | 60S ribosomal protein L28 | 0.714 |
| GB41142 | probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase-like isoform X2 | 0.712 |
| GB51359 | 60S ribosomal protein L27a isoform X1 | 0.710 |
| GB50519 | transmembrane emp24 domain-containing protein eca-like | 0.710 |
| GB47638 | ER membrane protein complex subunit 3-like | 0.705 |
| GB51046 | probable signal peptidase complex subunit 2-like | 0.701 |
| GB54973 | selT-like protein-like isoform 1 | 0.693 |
| GB44661 | intracellular protein transport protein USO1 isoform X9 | -0.691 |
| GB53953 | mitochondrial coenzyme A transporter SLC25A42-like isoformX1 | -0.688 |
| GB48289 | uncharacterized protein LOC726292 isoform X1 | 0.688 |
| GB47808 | DEP domain-containing protein 5 isoform X4 | -0.685 |
| GB45937 | intracellular protein transport protein USO1 isoform X2 | -0.684 |
| GB53750 | UPF0454 protein C12orf49 homolog isoform X2 | 0.682 |
| GB50455 | ubiquitin-conjugating enzyme E2-17 kDa-like | 0.676 |
| GB42356 | arginine-glutamic acid dipeptide repeats protein-like | -0.675 |
| GB51072 | 40S ribosomal protein S4-like isoform 1 | 0.669 |
| GB55268 | 43 kDa receptor-associated protein of the synapse homolog isoform X3 | -0.669 |
| GB43086 | uncharacterized protein LOC726486 | 0.668 |
| 102655259 | 5-methylcytosine rRNA methyltransferase NSUN4-like isoform X1 | -0.667 |
| GB55639 | 40S ribosomal protein S3 | 0.666 |
| GB41159 | bifunctional dihydrofolate reductase-thymidylate synthase | 0.665 |
| GB42354 | ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial-like isoform X4 | -0.663 |
| GB42696 | 60S ribosomal protein L35a isoform X3 | 0.656 |
| GB42088 | 40S ribosomal protein S29-like isoform X2 | 0.652 |
| GB53948 | uncharacterized protein LOC410057 isoform X1 | -0.651 |
| GB47553 | electron transfer flavoprotein subunit alpha, mitochondrial-like isoform 1 | 0.650 |
| 100577163 | slit homolog 2 protein-like | 0.650 |
| GB52627 | protein pigeon-like | -0.650 |
| GB54020 | apolipoprotein D-like | 0.650 |
| 102655440 | uncharacterized protein LOC102655440 | -0.649 |
| GB51009 | T-complex protein 1 subunit delta-like isoform 1 | 0.649 |
| GB49583 | 40S ribosomal protein S14 | 0.647 |
| GB41039 | 60S ribosomal protein L17 isoform 1 | 0.647 |
| GB46627 | paraplegin-like | -0.645 |
| GB54174 | E3 ubiquitin-protein ligase RING1 isoform 1 | 0.643 |
| GB41240 | aquaporin AQPAn.G-like isoform X3 | -0.641 |
| GB51440 | proteoglycan 4-like | -0.641 |
| GB45433 | small ribonucleoprotein particle protein B | 0.638 |
| GB51603 | peptidyl-alpha-hydroxyglycine alpha-amidating lyase 1-like isoform X5 | -0.638 |
| 100191002 | ribosomal protein L41 | 0.638 |
| GB43989 | serine-threonine kinase receptor-associated protein-like | 0.637 |
| GB53799 | proteasome subunit alpha type-2 | 0.636 |
| GB43141 | uncharacterized protein LOC413428 | -0.636 |
| GB44999 | chascon-like | -0.633 |
| GB49154 | bcl-2-related ovarian killer protein homolog A | 0.632 |
| GB50189 | epsilon-sarcoglycan | -0.630 |
| GB41150 | 40S ribosomal protein S2 isoform 2 | 0.628 |
| GB50917 | 60S acidic ribosomal protein P1 | 0.627 |
| GB48201 | 39S ribosomal protein L53, mitochondrial | 0.627 |
| GB44575 | ankyrin repeat and zinc finger domain-containing protein 1-like isoform X1 | -0.626 |
| GB46776 | 40S ribosomal protein S11 isoform X1 | 0.624 |
| GB49789 | 28S ribosomal protein S29, mitochondrial isoformX1 | -0.621 |
| GB46750 | 40S ribosomal protein S16 | 0.620 |
| GB44749 | 60S ribosomal protein L9 | 0.618 |
| GB44931 | evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial-like | -0.613 |
| GB46845 | 60S ribosomal protein L37a | 0.611 |
| GB43379 | membrane-bound transcription factor site-2 protease-like | 0.609 |
| GB45369 | receptor of activated protein kinase C 1, transcript variant X3 | 0.606 |
| GB52698 | synaptobrevin-like isoformX1 | 0.605 |
| 724829 | immediate early response 3-interacting protein 1-like isoform X1 | 0.605 |
| GB49536 | gamma-secretase subunit Aph-1 | 0.604 |
| GB55628 | probable RNA-binding protein EIF1AD-like isoform X1 | 0.603 |
| GB50832 | THO complex subunit 4-like | 0.602 |
| GB50929 | mitochondrial import receptor subunit TOM40 homolog 1-like isoform 1 | 0.601 |
| GB51065 | 40S ribosomal protein S10-like isoform 1 | 0.601 |
| GB54984 | chromatin complexes subunit BAP18-like isoform X1 | 0.600 |
| GB43180 | minor histocompatibility antigen H13-like | 0.597 |
| GB49365 | gamma-secretase subunit pen-2 isoform 1 | 0.595 |
| GB51543 | 60S ribosomal protein L13a isoform 2 | 0.594 |
| GB54341 | RNA-binding protein 33-like | -0.594 |
| 102655912 | L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase-like | 0.593 |
| GB53420 | uncharacterized protein LOC100576355 isoformX2 | -0.591 |
| GB48370 | ATP-binding cassette sub-family B member 7, mitochondrial isoform X1 | -0.591 |
| 726369 | peptidyl-tRNA hydrolase 2, mitochondrial-like isoform 1 | -0.591 |
| GB46478 | tectonin beta-propeller repeat-containing protein isoform X1 | -0.591 |
| GB42736 | TM2 domain-containing protein CG10795-like | 0.588 |
| GB49087 | formin-binding protein 1 homolog isoform X7 | -0.588 |
| GB50753 | uncharacterized LOC408705 | -0.586 |
| GB46123 | endonuclease G, mitochondrial-like | -0.586 |
| GB48574 | thioredoxin-2 isoform 1 | 0.586 |
| GB43232 | transmembrane protein 222-like isoform 1 | 0.586 |
| GB45285 | eukaryotic translation initiation factor 3 subunit F-like | 0.584 |
| GB44631 | uroporphyrinogen-III synthase-like | 0.582 |
| GB49994 | 60S ribosomal protein L26 | 0.581 |
| GB52563 | ATP-dependent helicase brm | -0.579 |
| GB54723 | uncharacterized protein LOC726790 isoform X1 | -0.578 |
| GB53668 | translocator protein-like | 0.577 |
| GB45374 | 40S ribosomal protein S23-like | 0.576 |
| GB46984 | ribonuclease UK114-like isoform 1 | 0.572 |
| 102655352 | uncharacterized protein LOC102655352 | -0.572 |
| GB45037 | beta-lactamase-like protein 2-like isoform X2 | -0.572 |
| GB52107 | tubulin alpha-1 chain-like | 0.571 |
| GB54139 | flocculation protein FLO11-like | -0.565 |
| 410017 | protein OPI10 homolog | 0.564 |
| GB49177 | 60S ribosomal protein L27 isoform X2 | 0.557 |
| GB54221 | transmembrane protein 50A-like | 0.556 |
| GB54979 | 60S ribosomal protein L21 | 0.555 |
| GB48111 | proteasome subunit beta type-1 | 0.552 |
| GB48745 | 5’-nucleotidase domain-containing protein 3-like | -0.550 |
| GB47079 | hexokinase type 2-like isoform X3 | -0.549 |
| GB47441 | V-type proton ATPase 21 kDa proteolipid subunit-like | 0.549 |
| GB41207 | 26S proteasome non-ATPase regulatory subunit 14 | 0.549 |
| GB50274 | transitional endoplasmic reticulum ATPase TER94 | 0.544 |
| GB51683 | annexin-B9-like isoform X1 | 0.544 |
| GB54952 | proteasome subunit alpha type-1-like | 0.543 |
| GB52253 | protein PRRC2C-like isoform X2 | -0.542 |
| GB41648 | protein chibby homolog 1-like | 0.542 |
| GB41363 | 26S protease regulatory subunit 6B isoform 1 | 0.541 |
| GB53247 | transmembrane emp24 domain-containing protein-like | 0.541 |
| GB48983 | RING finger protein 121-like isoform X3 | 0.541 |
| GB50873 | 60S ribosomal protein L30 isoform 1 | 0.540 |
| GB54255 | uncharacterized protein LOC551488 | 0.540 |
| GB48810 | 60S ribosomal protein L8 | 0.537 |
| GB41894 | uncharacterized protein LOC411277 isoform X28 | -0.536 |
| GB49021 | cuticular protein precursor | 0.536 |
| GB50131 | phosphatidate phosphatase PPAPDC1A-like isoform X2 | 0.535 |
| GB41811 | filaggrin-like isoform X3 | -0.534 |
| GB51484 | protein mago nashi | 0.528 |
| GB46705 | muscle M-line assembly protein unc-89 isoform X5 | -0.526 |
| GB45978 | dynein light chain Tctex-type isoform X2 | 0.526 |
| GB43449 | signal recognition particle 9 kDa protein | 0.526 |
| GB48150 | actin-related protein 2/3 complex subunit 1A | 0.525 |
| GB54854 | proteasome maturation protein-like | 0.523 |
| GB51545 | dystrophin, isoforms A/C/F/G/H-like | -0.523 |
| GB49095 | high affinity copper uptake protein 1-like isoformX1 | 0.523 |
| GB43638 | protein enhancer of sevenless 2B | 0.522 |
| GB51994 | proteasome subunit beta type-6-like | 0.520 |
| GB53194 | 60S ribosomal protein L14 isoform X2 | 0.519 |
| 102656618 | uncharacterized protein LOC102656618 isoform X1 | -0.518 |
| GB40539 | 40S ribosomal protein S20 | 0.518 |
| GB41631 | 60S ribosomal protein L34 isoform X2 | 0.518 |
| GB43938 | cytosolic endo-beta-N-acetylglucosaminidase-like isoform X4 | 0.516 |
| GB45878 | tRNA-dihydrouridine(16/17) synthase [NAD(P)(+)]-like isoform X3 | -0.516 |
| GB44039 | malate dehydrogenase, cytoplasmic-like isoform 1 | 0.513 |
| GB55781 | LOW QUALITY PROTEIN: uncharacterized protein LOC551170 | -0.513 |
| GB45526 | eukaryotic translation initiation factor 6 isoform 1 | 0.510 |
| GB52789 | 60S ribosomal protein L22 isoform 1 | 0.507 |
| GB53626 | myotrophin-like isoform 2 | 0.507 |
| GB49364 | splicing factor U2af 38 kDa subunit | 0.505 |
| GB44984 | U5 small nuclear ribonucleoprotein 40 kDa protein-like isoform X1 | 0.504 |
| GB50271 | zinc transporter 1-like | 0.503 |
| GB49377 | 40S ribosomal protein S3a | 0.501 |
| GB50874 | transcription factor Ken 2 | -0.501 |
| GB44147 | 60S ribosomal protein L15 | 0.498 |
| GB46141 | LOW QUALITY PROTEIN: vacuolar protein sorting-associated protein 29-like | 0.494 |
| GB51963 | mitochondrial ribonuclease P protein 1 homolog | -0.491 |
| GB55901 | ribosome biogenesis protein NSA2 homolog isoform X1 | 0.488 |
| GB42036 | protein SEC13 homolog isoform X2 | 0.485 |
| GB40877 | translocon-associated protein subunit delta | 0.485 |
| GB44205 | proteasome subunit beta type-5-like | 0.484 |
| GB54151 | uncharacterized protein LOC408463 isoform X12 | -0.484 |
| GB54590 | polyadenylate-binding protein 1-like isoform X2 | 0.483 |
| GB41157 | RPII140-upstream gene protein-like | -0.481 |
| GB48423 | small nuclear ribonucleoprotein F isoform 2 | 0.480 |
| GB49608 | protein angel-like isoform X1 | -0.475 |
| GB49812 | RING-box protein 1A isoform X1 | 0.461 |
| GB43697 | mediator of RNA polymerase II transcription subunit 16 isoform X3 | 0.457 |
| GB41553 | Golgi phosphoprotein 3 homolog rotini-like isoform X1 | 0.455 |
| GB43548 | 40S ribosomal protein SA | 0.449 |
| GB45181 | probable Bax inhibitor 1 | 0.447 |
| GB53086 | alcohol dehydrogenase class-3 isoform X2 | 0.446 |
| GB41724 | uncharacterized protein LOC727081 | -0.446 |
| GB54533 | protein unc-13 homolog D isoform X5 | -0.445 |
| GB40882 | 40S ribosomal protein S13 isoform X1 | 0.445 |
| GB50230 | V-type proton ATPase subunit e 2-like | 0.445 |
| 102654691 | protein translation factor SUI1 homolog | 0.445 |
| GB51787 | myosin light chain alkali-like isoform X5 | 0.444 |
| GB41908 | PERQ amino acid-rich with GYF domain-containing protein CG11148-like isoform X3 | -0.443 |
| GB53138 | inorganic pyrophosphatase-like | 0.437 |
| GB48250 | putative gamma-glutamylcyclotransferase CG2811-like isoform X4 | 0.436 |
| GB46763 | excitatory amino acid transporter 3 | 0.436 |
| GB53415 | WW domain-binding protein 2-like isoform X1 | 0.432 |
| GB47606 | ER membrane protein complex subunit 4-like isoform 1 | 0.431 |
| GB42675 | adenylate cyclase type 2-like | -0.430 |
| GB44312 | hydroxyacylglutathione hydrolase, mitochondrial-like isoform X2 | 0.428 |
| 102654127 | neurochondrin homolog | -0.423 |
| GB47938 | uncharacterized protein LOC412825 isoform X1 | -0.421 |
| GB55056 | spermatogenesis-associated protein 20 isoform X2 | -0.420 |
| GB41084 | 60S ribosomal protein L38 | 0.412 |
| GB47810 | regulator of gene activity protein isoform X3 | 0.410 |
| GB40946 | serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform-like isoform X1 | 0.409 |
| GB42780 | CCHC-type zinc finger protein CG3800-like isoform X3 | 0.409 |
| GB43229 | GTP-binding nuclear protein Ran isoform X1 | 0.407 |
| GB50244 | NHL repeat-containing protein 2 isoform X4 | -0.406 |
| GB45684 | protein spire-like isoform X4 | -0.406 |
| GB52073 | probable citrate synthase 1, mitochondrial-like | -0.402 |
| GB45856 | protein GPR107-like isoform X4 | 0.401 |
| GB47542 | eukaryotic translation initiation factor 3 subunit J isoform 1 | 0.399 |
| GB53243 | LOW QUALITY PROTEIN: probable phosphorylase b kinase regulatory subunit beta-like | -0.395 |
| GB55892 | glutamate-rich WD repeat-containing protein 1-like | 0.395 |
| GB43105 | casein kinase II subunit alpha isoform X6 | 0.390 |
| GB43537 | probable 28S ribosomal protein S16, mitochondrial | 0.388 |
| GB44496 | probable serine incorporator isoformX1 | 0.376 |
| GB43855 | LOW QUALITY PROTEIN: coatomer subunit beta’ | 0.374 |
| GB40073 | COP9 signalosome complex subunit 8-like | 0.373 |
| GB47100 | putative glutamate synthase [NADPH]-like isoform X4 | -0.372 |
| GB42786 | microtubule-associated protein RP/EB family member 1-like isoform X4 | 0.372 |
| GB54789 | GMP synthase [glutamine-hydrolyzing] | 0.371 |
| GB40767 | phosphoglycolate phosphatase-like | -0.370 |
| GB52212 | polyubiquitin-A-like isoform X2 | 0.370 |
| GB52256 | 60S ribosomal protein L5 | 0.370 |
| GB50925 | prostaglandin E synthase 2-like | -0.360 |
| GB53725 | splicing factor 3B subunit 1-like isoform X2 | -0.355 |
| GB44870 | zinc finger protein 706-like isoform X3 | 0.355 |
| GB45375 | rhomboid-7 isoform X1 | 0.346 |
| GB45044 | uncharacterized protein LOC409396 isoform X5 | -0.334 |
| GB50909 | dual 3’,5’-cyclic-AMP and -GMP phosphodiesterase 11-like, transcript variant X4 | -0.333 |
| GB44907 | myeloid leukemia factor isoform X3 | 0.331 |
| GB44333 | flocculation protein FLO11-like isoform X1 | -0.322 |
| GB46562 | 40S ribosomal protein S24-like isoform X2 | 0.321 |
| GB45017 | RNA pseudouridylate synthase domain-containing protein 2-like isoform X3 | -0.317 |
| GB48312 | pre-mRNA-splicing factor RBM22-like | 0.316 |
| GB47103 | elongation factor 1-beta’ | 0.310 |
| GB48207 | proteasomal ubiquitin receptor ADRM1 homolog isoform X1 | 0.270 |
| GB40887 | V-type proton ATPase subunit E isoform 3 | 0.266 |
| GB41152 | uncharacterized protein C6orf106 homolog | 0.213 |
| GB45678 | 1-acylglycerol-3-phosphate O-acyltransferase ABHD5-like isoform X1 | -0.192 |
| GB44576 | ester hydrolase C11orf54 homolog | 0.189 |
Bombus terrestris
Table S3: The single significantly differentially expressed gene (EBseq; FDR-corrected posterior probability of differential expression p < 0.05) in Bombus terrestris. Positive fold change values indicate higher expression in the control, while negative values indicate higher expression in the queen pheromone treatment. The second and third columns give the best BLAST hit for this gene in A. mellifera plus the name of the A. mellifera putative ortholog.
saveRDS(bombus.de, file = "supplement/tab_S3.rds")
kable(bombus.de, "html") %>%
kable_styling()| Gene | Apis BLAST | Name | Log2 FC |
|---|---|---|---|
| 100648170 | GB48391 | mucin-2-like | 1.071 |
Lasius flavus
Table S4: List of the 290 significantly differentially expressed genes (EBseq; FDR-corrected posterior probability of differential expression p < 0.05) in Lasius flavus, listed in order of fold change in gene expression on a Log\(_2\) scale. Positive fold change values indicate higher expression in the control, while negative values indicate higher expression in the queen pheromone treatment. The second and third columns give the best BLAST hit for this gene in A. mellifera plus the name of the A. mellifera putative ortholog.
saveRDS(flavus.de, file = "supplement/tab_S4.rds")
kable.table(flavus.de)| Gene | Apis BLAST | Name | Log2 FC |
|---|---|---|---|
| TRINITY_DN19074_c0_g2 | GB43902 | hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like | 7.514 |
| TRINITY_DN2701_c0_g1 | GB52729 | aspartate–tRNA ligase, cytoplasmic | 7.481 |
| TRINITY_DN14108_c0_g2 | 7.468 | ||
| TRINITY_DN36041_c0_g1 | 6.629 | ||
| TRINITY_DN13621_c0_g1 | 6.549 | ||
| TRINITY_DN18780_c0_g2 | -6.440 | ||
| TRINITY_DN14430_c0_g1 | 6.278 | ||
| TRINITY_DN32671_c0_g2 | 6.121 | ||
| TRINITY_DN14910_c0_g1 | XP_016769216.1 | 5.800 | |
| TRINITY_DN19071_c0_g1 | -5.776 | ||
| TRINITY_DN5663_c0_g2 | XP_006568418.2 | -5.365 | |
| TRINITY_DN13527_c1_g3 | 5.259 | ||
| TRINITY_DN2506_c0_g2 | 5.133 | ||
| TRINITY_DN9902_c0_g2 | 5.069 | ||
| TRINITY_DN6503_c0_g3 | 551397 | 28S ribosomal protein S18a, mitochondrial isoform 2 | 5.044 |
| TRINITY_DN10699_c0_g1 | 4.901 | ||
| TRINITY_DN5102_c0_g1 | XP_016771437.1 | 4.901 | |
| TRINITY_DN6994_c0_g1 | 4.859 | ||
| TRINITY_DN13376_c3_g2 | 4.858 | ||
| TRINITY_DN9565_c0_g3 | GB45250 | uncharacterized protein LOC409595 | 4.811 |
| TRINITY_DN1013_c0_g1 | GB52059 | eukaryotic translation initiation factor 4H-like isoform X1 | 4.709 |
| TRINITY_DN11616_c0_g1 | 4.696 | ||
| TRINITY_DN1845_c0_g2 | GB52253 | protein PRRC2C-like isoform X2 | 4.658 |
| TRINITY_DN7242_c0_g1 | -4.646 | ||
| TRINITY_DN31547_c0_g3 | 4.613 | ||
| TRINITY_DN13376_c3_g3 | 4.569 | ||
| TRINITY_DN6298_c0_g3 | 4.553 | ||
| TRINITY_DN2720_c0_g2 | -4.522 | ||
| TRINITY_DN2583_c0_g2 | 4.460 | ||
| TRINITY_DN24980_c0_g3 | -4.393 | ||
| TRINITY_DN12959_c0_g3 | 4.390 | ||
| TRINITY_DN4813_c0_g1 | 4.322 | ||
| TRINITY_DN6331_c0_g2 | 4.303 | ||
| TRINITY_DN10925_c0_g1 | 4.278 | ||
| TRINITY_DN14174_c1_g3 | 4.233 | ||
| TRINITY_DN23574_c0_g3 | 4.173 | ||
| TRINITY_DN6054_c0_g1 | XP_016771772.1 | 4.143 | |
| TRINITY_DN32287_c0_g1 | 4.133 | ||
| TRINITY_DN1060_c0_g1 | 4.115 | ||
| TRINITY_DN6965_c0_g3 | 4.097 | ||
| TRINITY_DN5087_c0_g1 | XP_003249576.2 | 4.069 | |
| TRINITY_DN10904_c0_g1 | 4.050 | ||
| TRINITY_DN4900_c0_g1 | 4.049 | ||
| TRINITY_DN14065_c0_g2 | 4.015 | ||
| TRINITY_DN2102_c0_g6 | GB40389 | profilin | 3.751 |
| TRINITY_DN12195_c1_g3 | 102656074 | reticulon-4-like isoform X6 | 3.710 |
| TRINITY_DN2302_c0_g2 | 3.676 | ||
| TRINITY_DN9563_c1_g3 | XP_016770117.1 | 3.347 | |
| TRINITY_DN11562_c2_g1 | XP_016769630.1 | 3.211 | |
| TRINITY_DN6503_c0_g2 | 551397 | 28S ribosomal protein S18a, mitochondrial isoform 2 | 3.210 |
| TRINITY_DN3428_c0_g1 | GB53155 | maternal embryonic leucine zipper kinase-like | 3.168 |
| TRINITY_DN11833_c0_g2 | 3.162 | ||
| TRINITY_DN1150_c0_g2 | 3.151 | ||
| TRINITY_DN14156_c9_g2 | XP_016769763.1 | 3.146 | |
| TRINITY_DN7447_c0_g1 | GB10293 | aubergine | 3.125 |
| TRINITY_DN12563_c0_g2 | XP_016768561.1 | 3.097 | |
| TRINITY_DN19328_c0_g2 | GB49105 | ecdysteroid-regulated gene E74 isoform X10 | 2.921 |
| TRINITY_DN19071_c0_g5 | -2.920 | ||
| TRINITY_DN3355_c0_g1 | XP_016772030.1 | 2.912 | |
| TRINITY_DN1453_c0_g2 | 2.891 | ||
| TRINITY_DN34334_c0_g1 | -2.831 | ||
| TRINITY_DN9365_c0_g1 | 2.823 | ||
| TRINITY_DN2907_c0_g1 | GB52114 | protein trachealess-like isoform X7 | 2.821 |
| TRINITY_DN29060_c0_g1 | 2.784 | ||
| TRINITY_DN5450_c0_g3 | 2.778 | ||
| TRINITY_DN6679_c1_g1 | -2.777 | ||
| TRINITY_DN13106_c0_g1 | -2.766 | ||
| TRINITY_DN6249_c0_g2 | 2.762 | ||
| TRINITY_DN12570_c0_g2 | XP_016772046.1 | 2.752 | |
| TRINITY_DN29060_c0_g2 | -2.748 | ||
| TRINITY_DN1453_c0_g1 | 2.718 | ||
| TRINITY_DN4551_c0_g1 | 2.665 | ||
| TRINITY_DN5934_c0_g2 | 2.664 | ||
| TRINITY_DN21319_c0_g1 | 2.647 | ||
| TRINITY_DN16415_c0_g2 | 2.644 | ||
| TRINITY_DN6639_c0_g2 | GB51740 | CD63 antigen | 2.628 |
| TRINITY_DN8686_c0_g6 | 2.525 | ||
| TRINITY_DN27412_c0_g1 | 2.511 | ||
| TRINITY_DN7556_c0_g1 | 2.479 | ||
| TRINITY_DN12097_c3_g11 | 2.467 | ||
| TRINITY_DN19324_c0_g1 | 2.464 | ||
| TRINITY_DN5257_c0_g2 | GB51614 | probable methylthioribulose-1-phosphate dehydratase-like | 2.451 |
| TRINITY_DN3033_c0_g1 | GB47735 | endonuclease III-like protein 1-like | 2.441 |
| TRINITY_DN23065_c0_g1 | 2.434 | ||
| TRINITY_DN30835_c0_g2 | 2.337 | ||
| TRINITY_DN30278_c0_g7 | 2.328 | ||
| TRINITY_DN13221_c0_g7 | 2.244 | ||
| TRINITY_DN13083_c0_g1 | 2.202 | ||
| TRINITY_DN12097_c3_g6 | 2.167 | ||
| TRINITY_DN6372_c0_g3 | 2.154 | ||
| TRINITY_DN13237_c2_g6 | -2.137 | ||
| TRINITY_DN3870_c0_g2 | 2.108 | ||
| TRINITY_DN7016_c0_g2 | GB47843 | uncharacterized protein LOC100576559 isoform X2 | 1.972 |
| TRINITY_DN12195_c1_g4 | 102656074 | reticulon-4-like isoform X6 | -1.727 |
| TRINITY_DN15459_c0_g1 | 1.644 | ||
| TRINITY_DN7587_c0_g2 | GB52590 | fatty acid synthase-like isoform 1 | -1.363 |
| TRINITY_DN3266_c0_g1 | 1.320 | ||
| TRINITY_DN5353_c0_g1 | GB43825 | lysosomal aspartic protease | 1.297 |
| TRINITY_DN7865_c0_g1 | XP_016770671.1 | 1.257 | |
| TRINITY_DN13667_c2_g1 | 1.226 | ||
| TRINITY_DN9568_c0_g3 | -1.068 | ||
| TRINITY_DN8668_c0_g1 | GB52590 | fatty acid synthase-like isoform 1 | -1.025 |
| TRINITY_DN8075_c0_g1 | -1.023 | ||
| TRINITY_DN8944_c1_g2 | -0.955 | ||
| TRINITY_DN13195_c1_g2 | 0.950 | ||
| TRINITY_DN11726_c1_g1 | GB52590 | fatty acid synthase-like isoform 1 | -0.932 |
| TRINITY_DN14247_c8_g2 | GB52590 | fatty acid synthase-like isoform 1 | -0.926 |
| TRINITY_DN61_c0_g1 | GB45775 | pancreatic triacylglycerol lipase-like isoform X2 | -0.888 |
| TRINITY_DN2623_c0_g1 | GB43825 | lysosomal aspartic protease | 0.862 |
| TRINITY_DN12575_c0_g1 | GB55263 | putative fatty acyl-CoA reductase CG5065-like | -0.832 |
| TRINITY_DN14020_c0_g1 | GB46188 | trichohyalin-like isoform X1 | 0.728 |
| TRINITY_DN13013_c0_g1 | XP_016768441.1 | 0.710 | |
| TRINITY_DN12756_c2_g1 | 0.691 | ||
| TRINITY_DN9649_c0_g1 | NP_001305411.1 | 0.685 | |
| TRINITY_DN13574_c1_g2 | GB40681 | elongation of very long chain fatty acids protein 1-like | -0.679 |
| TRINITY_DN9287_c0_g1 | XP_016768964.1 | 0.671 | |
| TRINITY_DN13318_c0_g1 | GB46888 | alpha-methylacyl-CoA racemase-like | -0.612 |
| TRINITY_DN3111_c0_g1 | 0.588 | ||
| TRINITY_DN13226_c0_g1 | GB47475 | protein lethal(2)essential for life-like isoform 1 | 0.586 |
| TRINITY_DN5134_c0_g1 | XP_016770229.1 | 0.578 | |
| TRINITY_DN12647_c0_g1 | XP_016773029.1 | -0.572 | |
| TRINITY_DN14059_c0_g1 | XP_016768888.1 | 0.567 | |
| TRINITY_DN13252_c1_g1 | GB50415 | diacylglycerol kinase theta-like isoform X7 | 0.559 |
| TRINITY_DN13742_c0_g1 | GB46657 | galactokinase-like | -0.518 |
| TRINITY_DN13982_c0_g1 | XP_016769706.1 | -0.517 | |
| TRINITY_DN32324_c0_g1 | 102656101 | uncharacterized protein LOC102656101 | 0.517 |
| TRINITY_DN12496_c0_g1 | GB54423 | uncharacterized protein LOC551958 | 0.511 |
| TRINITY_DN11328_c0_g1 | GB51479 | ras guanine nucleotide exchange factor P-like isoform X3 | 0.509 |
| TRINITY_DN6523_c0_g1 | GB40976 | heat shock protein 90 | -0.504 |
| TRINITY_DN12344_c0_g1 | 0.503 | ||
| TRINITY_DN15124_c0_g1 | 0.500 | ||
| TRINITY_DN12742_c1_g1 | XP_016767680.1 | 0.490 | |
| TRINITY_DN11193_c0_g1 | GB53045 | ATP-binding cassette sub-family G member 1-like isoform X1 | -0.481 |
| TRINITY_DN10448_c0_g2 | GB41603 | PTB domain-containing adapter protein ced-6 isoform X2 | -0.474 |
| TRINITY_DN1154_c0_g1 | 0.474 | ||
| TRINITY_DN14164_c3_g1 | GB55490 | uncharacterized protein LOC410793 | -0.473 |
| TRINITY_DN14136_c5_g1 | GB55016 | quinone oxidoreductase-like isoform X2 | 0.465 |
| TRINITY_DN9952_c0_g1 | GB43823 | chemosensory protein 1 precursor | 0.462 |
| TRINITY_DN8450_c0_g1 | GB46286 | zinc carboxypeptidase A 1-like isoform X1 | 0.454 |
| TRINITY_DN12807_c0_g1 | XP_016769434.1 | 0.450 | |
| TRINITY_DN13250_c5_g1 | GB45937 | intracellular protein transport protein USO1 isoform X2 | 0.445 |
| TRINITY_DN6544_c0_g1 | GB52074 | 6-phosphogluconate dehydrogenase, decarboxylating | -0.445 |
| TRINITY_DN13581_c2_g2 | GB42792 | uncharacterized protein LOC409805 isoform X3 | 0.444 |
| TRINITY_DN36181_c0_g1 | 0.442 | ||
| TRINITY_DN2719_c0_g1 | GB54446 | arginine kinase isoform X2 | 0.441 |
| TRINITY_DN13519_c0_g1 | GB42797 | protein takeout-like | 0.440 |
| TRINITY_DN7060_c0_g1 | GB49607 | lysosome-associated membrane glycoprotein 1-like isoform 2 | -0.438 |
| TRINITY_DN1392_c0_g1 | GB44205 | proteasome subunit beta type-5-like | -0.434 |
| TRINITY_DN10466_c0_g1 | GB44431 | 26S protease regulatory subunit 4 isoform 1 | -0.428 |
| TRINITY_DN13148_c0_g1 | GB44213 | filamin-like | 0.426 |
| TRINITY_DN13702_c3_g1 | XP_016769732.1 | 0.423 | |
| TRINITY_DN12417_c0_g2 | GB45456 | flocculation protein FLO11-like isoform X2 | 0.422 |
| TRINITY_DN23399_c0_g1 | 0.417 | ||
| TRINITY_DN11737_c0_g1 | XP_016766478.1 | -0.416 | |
| TRINITY_DN12348_c0_g1 | GB44703 | proteasome activator complex subunit 4-like | -0.415 |
| TRINITY_DN13581_c1_g3 | XP_016767189.1 | 0.406 | |
| TRINITY_DN11774_c0_g1 | XP_016772498.1 | 0.387 | |
| TRINITY_DN3048_c0_g1 | GB40770 | dehydrogenase/reductase SDR family member 11-like isoform X2 | 0.387 |
| TRINITY_DN13700_c5_g3 | GB42840 | leukocyte receptor cluster member 8 homolog isoform X4 | 0.387 |
| TRINITY_DN13455_c0_g1 | GB45128 | trifunctional enzyme subunit alpha, mitochondrial-like | -0.380 |
| TRINITY_DN14019_c2_g1 | 409060 | neurofilament heavy polypeptide-like isoform X2 | 0.377 |
| TRINITY_DN11786_c1_g1 | GB51214 | troponin T, skeletal muscle | 0.374 |
| TRINITY_DN9982_c0_g1 | GB51787 | myosin light chain alkali-like isoform X5 | 0.372 |
| TRINITY_DN27322_c0_g1 | GB40866 | heat shock protein cognate 4 | -0.369 |
| TRINITY_DN5956_c0_g1 | 0.366 | ||
| TRINITY_DN6208_c0_g1 | GB49757 | fatty acid binding protein | 0.359 |
| TRINITY_DN11594_c0_g1 | GB52643 | poly(U)-specific endoribonuclease homolog | 0.355 |
| TRINITY_DN9687_c0_g1 | 0.350 | ||
| TRINITY_DN14119_c3_g1 | 726668 | PDZ and LIM domain protein 3 isoform X7 | 0.348 |
| TRINITY_DN5256_c0_g1 | 0.345 | ||
| TRINITY_DN10396_c0_g1 | GB42607 | cytochrome b5-like isoform X1 | -0.344 |
| TRINITY_DN5623_c0_g1 | GB54817 | muscle-specific protein 20 | 0.343 |
| TRINITY_DN10620_c0_g1 | GB42732 | long-chain-fatty-acid–CoA ligase 3-like isoform X2 | -0.325 |
| TRINITY_DN12788_c0_g1 | XP_016771468.1 | 0.325 | |
| TRINITY_DN12757_c0_g1 | GB55610 | MOSC domain-containing protein 2, mitochondrial-like | 0.324 |
| TRINITY_DN10923_c0_g1 | GB40141 | venom serine carboxypeptidase | -0.321 |
| TRINITY_DN10232_c0_g1 | -0.319 | ||
| TRINITY_DN12105_c0_g1 | XP_016768441.1 | 0.318 | |
| TRINITY_DN7549_c0_g1 | XP_016768456.1 | 0.315 | |
| TRINITY_DN3036_c0_g1 | GB52326 | chemosensory protein 4 precursor | 0.313 |
| TRINITY_DN12756_c2_g4 | XP_016770894.1 | 0.311 | |
| TRINITY_DN14002_c3_g1 | XP_016770982.1 | 0.307 | |
| TRINITY_DN27284_c0_g1 | GB50274 | transitional endoplasmic reticulum ATPase TER94 | -0.306 |
| TRINITY_DN12138_c0_g1 | GB47306 | sulfhydryl oxidase 1-like | 0.305 |
| TRINITY_DN13865_c0_g1 | GB47963 | probable E3 ubiquitin-protein ligase HERC4-like isoform X3 | 0.302 |
| TRINITY_DN8423_c0_g1 | XP_016768214.1 | -0.298 | |
| TRINITY_DN27569_c0_g1 | GB52736 | ATP synthase subunit beta, mitochondrial isoform X1 | 0.297 |
| TRINITY_DN14286_c2_g1 | GB54861 | LOW QUALITY PROTEIN: counting factor associated protein D-like | -0.294 |
| TRINITY_DN14128_c1_g1 | XP_006568818.2 | 0.291 | |
| TRINITY_DN11359_c0_g1 | XP_016768872.1 | -0.287 | |
| TRINITY_DN9072_c0_g1 | XP_016768321.1 | -0.287 | |
| TRINITY_DN28113_c0_g1 | -0.285 | ||
| TRINITY_DN10911_c0_g1 | XP_016770213.1 | -0.277 | |
| TRINITY_DN12726_c3_g1 | GB42787 | dentin sialophosphoprotein-like isoform X4 | -0.274 |
| TRINITY_DN6072_c0_g1 | XP_016771431.1 | -0.273 | |
| TRINITY_DN13789_c0_g2 | XP_016767109.1 | 0.273 | |
| TRINITY_DN14037_c0_g1 | XP_016767155.1 | 0.272 | |
| TRINITY_DN13738_c0_g1 | XP_016772667.1 | 0.271 | |
| TRINITY_DN993_c0_g3 | 0.270 | ||
| TRINITY_DN7531_c0_g3 | GB51710 | eukaryotic initiation factor 4A-like isoformX2 | 0.260 |
| TRINITY_DN11232_c0_g1 | GB40240 | myosin regulatory light chain 2 | 0.258 |
| TRINITY_DN12074_c1_g1 | GB47462 | protein disulfide-isomerase A3 isoform 2 | -0.256 |
| TRINITY_DN11684_c0_g1 | GB55537 | transketolase isoform 1 | -0.248 |
| TRINITY_DN14138_c2_g1 | GB48850 | fatty-acid amide hydrolase 2-B-like | -0.248 |
| TRINITY_DN13963_c1_g1 | XP_016768450.1 | 0.247 | |
| TRINITY_DN12180_c0_g1 | XP_016772046.1 | 0.246 | |
| TRINITY_DN13233_c1_g1 | XP_016768217.1 | -0.244 | |
| TRINITY_DN12080_c1_g1 | XP_016769481.1 | -0.241 | |
| TRINITY_DN11171_c0_g1 | GB42468 | phospholipase B1, membrane-associated-like isoform X1 | -0.239 |
| TRINITY_DN13982_c0_g3 | XP_016769706.1 | -0.237 | |
| TRINITY_DN14752_c0_g1 | GB50123 | myophilin-like | 0.226 |
| TRINITY_DN8270_c0_g1 | GB43276 | aminopeptidase N-like isoform X1 | 0.226 |
| TRINITY_DN12844_c1_g1 | GB47885 | probable cytochrome P450 304a1 | -0.224 |
| TRINITY_DN11885_c1_g1 | GB55598 | troponin I isoform X23 | 0.218 |
| TRINITY_DN8742_c0_g1 | XP_016767150.1 | -0.216 | |
| TRINITY_DN14111_c0_g1 | GB46705 | muscle M-line assembly protein unc-89 isoform X5 | 0.213 |
| TRINITY_DN14002_c4_g1 | 0.212 | ||
| TRINITY_DN7802_c0_g1 | GB41358 | elongation factor 1-alpha | -0.210 |
| TRINITY_DN14006_c0_g1 | XP_016767101.1 | -0.208 | |
| TRINITY_DN8847_c0_g1 | GB46772 | very-long-chain enoyl-CoA reductase-like | -0.201 |
| TRINITY_DN14179_c1_g1 | GB40461 | calreticulin | -0.186 |
| TRINITY_DN7523_c0_g1 | -0.183 | ||
| TRINITY_DN12909_c0_g1 | XP_016770377.1 | 0.181 | |
| TRINITY_DN8174_c0_g1 | GB47880 | superoxide dismutase 1 | -0.175 |
| TRINITY_DN14199_c2_g1 | -0.173 | ||
| TRINITY_DN3676_c0_g2 | GB49773 | sequestosome-1 | 0.168 |
| TRINITY_DN3648_c0_g1 | GB45181 | probable Bax inhibitor 1 | -0.167 |
| TRINITY_DN9738_c0_g1 | GB44206 | death-associated protein 1-like | -0.167 |
| TRINITY_DN3697_c0_g1 | GB54368 | prostaglandin E synthase 3-like isoform X2 | -0.165 |
| TRINITY_DN13223_c0_g1 | GB54315 | uncharacterized protein LOC724126 | -0.157 |
| TRINITY_DN9957_c0_g1 | GB43831 | ATP-binding cassette sub-family D member 3-like | -0.157 |
| TRINITY_DN1533_c0_g1 | XP_392401.3 | -0.155 | |
| TRINITY_DN12307_c0_g1 | GB47029 | uncharacterized protein LOC724558 | -0.153 |
| TRINITY_DN13021_c0_g1 | XP_006571535.2 | -0.144 | |
| TRINITY_DN11571_c0_g2 | XP_001119981.3 | -0.138 | |
| TRINITY_DN14152_c0_g10 | XP_016771978.1 | 0.136 | |
| TRINITY_DN13002_c1_g1 | XP_016767675.1 | -0.131 | |
| TRINITY_DN13997_c1_g2 | XP_016771269.1 | 0.130 | |
| TRINITY_DN7931_c0_g1 | GB49321 | D-arabinitol dehydrogenase 1-like | -0.128 |
| TRINITY_DN12756_c2_g5 | XP_016770894.1 | -0.126 | |
| TRINITY_DN1575_c0_g1 | 0.124 | ||
| TRINITY_DN1616_c0_g1 | GB41545 | MD-2-related lipid-recognition protein-like | 0.120 |
| TRINITY_DN12630_c0_g1 | GB45258 | isocitrate dehydrogenase [NADP] cytoplasmic isoform 2 | -0.119 |
| TRINITY_DN14083_c3_g1 | GB55263 | putative fatty acyl-CoA reductase CG5065-like | -0.117 |
| TRINITY_DN4494_c0_g2 | GB47990 | tropomyosin-1-like | 0.116 |
| TRINITY_DN1524_c0_g1 | GB46920 | iron-sulfur cluster assembly enzyme ISCU, mitochondrial | 0.115 |
| TRINITY_DN13221_c0_g9 | XP_016769341.1 | 0.107 | |
| TRINITY_DN13959_c2_g1 | GB49688 | peroxidase isoformX2 | 0.104 |
| TRINITY_DN12634_c0_g1 | GB43575 | trehalase-like isoform X2 | -0.093 |
| TRINITY_DN13844_c1_g1 | XP_016767538.1 | -0.092 | |
| TRINITY_DN10322_c0_g1 | XP_016769014.1 | -0.088 | |
| TRINITY_DN13381_c0_g1 | GB51633 | protein HIRA homolog | 0.084 |
| TRINITY_DN12970_c0_g2 | GB44208 | WD repeat-containing protein 37-like isoform X4 | 0.083 |
| TRINITY_DN3647_c0_g1 | GB53550 | heat shock protein beta-1-like isoform X3 | -0.081 |
| TRINITY_DN8558_c0_g1 | GB46713 | translation elongation factor 2-like isoform 1 | -0.079 |
| TRINITY_DN11372_c0_g1 | GB53755 | juvenile hormone esterase precursor | 0.071 |
| TRINITY_DN6049_c0_g1 | 0.068 | ||
| TRINITY_DN10813_c0_g1 | GB51753 | uncharacterized protein LOC100576760 isoform X2 | -0.066 |
| TRINITY_DN10427_c0_g1 | GB51782 | carboxypeptidase Q-like isoform 1 | -0.065 |
| TRINITY_DN8685_c0_g4 | GB43825 | lysosomal aspartic protease | -0.065 |
| TRINITY_DN13047_c0_g1 | XP_016768229.1 | 0.061 | |
| TRINITY_DN6208_c0_g2 | GB49757 | fatty acid binding protein | -0.056 |
| TRINITY_DN13652_c0_g1 | GB42422 | ADP/ATP translocase | 0.054 |
| TRINITY_DN13884_c0_g2 | GB47405 | neutral alpha-glucosidase AB-like isoform 2 | 0.051 |
| TRINITY_DN13634_c0_g1 | GB45913 | protein lethal(2)essential for life-like | -0.049 |
| TRINITY_DN1723_c0_g1 | GB52324 | chemosensory protein 3 precursor | 0.048 |
| TRINITY_DN18895_c0_g1 | GB55581 | membrane-associated progesterone receptor component 1-like isoform 2 | -0.044 |
| TRINITY_DN8126_c0_g1 | GB40779 | transaldolase | -0.044 |
| TRINITY_DN10665_c0_g1 | GB42829 | juvenile hormone epoxide hydrolase 1 | -0.042 |
| TRINITY_DN8184_c0_g1 | 0.040 | ||
| TRINITY_DN11577_c0_g1 | GB55096 | NADP-dependent malic enzyme isoform X3 | 0.040 |
| TRINITY_DN18992_c0_g1 | XP_016772082.1 | 0.039 | |
| TRINITY_DN11030_c0_g1 | GB49240 | aldehyde dehydrogenase, mitochondrial isoform 1 | -0.038 |
| TRINITY_DN11459_c0_g1 | GB50598 | aldose reductase-like isoform 1 | -0.038 |
| TRINITY_DN13961_c1_g3 | GB52588 | conserved oligomeric Golgi complex subunit 7 | -0.037 |
| TRINITY_DN12872_c0_g1 | GB53333 | V-type proton ATPase catalytic subunit A-like isoform X3 | 0.035 |
| TRINITY_DN13271_c0_g1 | GB40312 | choline/ethanolamine kinase-like isoform X4 | 0.030 |
| TRINITY_DN13702_c8_g1 | GB47395 | uncharacterized protein CG7816-like | -0.030 |
| TRINITY_DN13400_c0_g1 | GB54421 | uncharacterized protein DDB_G0287625-like | 0.023 |
| TRINITY_DN10682_c0_g1 | GB50252 | GTP-binding protein SAR1b-like isoform X4 | -0.021 |
| TRINITY_DN8055_c0_g1 | -0.021 | ||
| TRINITY_DN9151_c0_g1 | GB45147 | clavesin-2-like | 0.021 |
| TRINITY_DN11885_c3_g1 | GB47880 | superoxide dismutase 1 | -0.019 |
| TRINITY_DN12979_c1_g1 | GB49347 | prostaglandin reductase 1-like | 0.013 |
| TRINITY_DN14085_c1_g1 | XP_016769919.1 | -0.011 | |
| TRINITY_DN11118_c1_g1 | GB44422 | uncharacterized protein LOC412543 isoform X3 | -0.010 |
| TRINITY_DN14002_c1_g1 | 0.004 | ||
| TRINITY_DN7306_c0_g2 | XP_016769332.1 | -0.004 | |
| TRINITY_DN13747_c0_g1 | XP_016769944.1 | -0.003 | |
| TRINITY_DN10790_c0_g1 | 0.000 |
Lasius niger
Table S5: List of the 135 significantly differentially expressed genes (EBseq; FDR-corrected posterior probability of differential expression p < 0.05) in Lasius niger, listed in order of fold change in gene expression on a Log\(_2\) scale. Positive fold change values indicate higher expression in the control, while negative values indicate higher expression in the queen pheromone treatment. The second and third columns give the best BLAST hit for this gene in A. mellifera plus the name of the A. mellifera putative ortholog.
saveRDS(niger.de, file = "supplement/tab_S5.rds")
kable.table(niger.de)| Gene | Apis BLAST | Name | Log2 FC |
|---|---|---|---|
| XLOC_001009 | 5.916 | ||
| RF55_9944 | GB55171 | major royal jelly protein 1 isoform X1 | 5.060 |
| RF55_873 | XP_016773511.1 | 4.638 | |
| XLOC_000784 | 3.626 | ||
| RF55_874 | XP_016766165.1 | 3.550 | |
| XLOC_016588 | 3.077 | ||
| RF55_9436 | 2.813 | ||
| RF55_3510 | GB53672 | failed axon connections isoform X2 | -2.471 |
| RF55_15864 | -2.439 | ||
| XLOC_020552 | 2.437 | ||
| RF55_783 | 2.206 | ||
| RF55_6001 | -2.150 | ||
| XLOC_013573 | 2.150 | ||
| RF55_4870 | XP_006563262.2 | -2.031 | |
| XLOC_022706 | -1.091 | ||
| RF55_7689 | XP_003250465.2 | -1.030 | |
| RF55_19841 | GB52590 | fatty acid synthase-like isoform 1 | -1.015 |
| RF55_21338 | GB52590 | fatty acid synthase-like isoform 1 | -0.923 |
| RF55_13568 | XP_006571191.2 | 0.882 | |
| RF55_2210 | GB49869 | microsomal triglyceride transfer protein large subunit isoform X1 | -0.879 |
| RF55_6639 | GB48784 | cytochrome c | 0.779 |
| RF55_15245 | GB43617 | uncharacterized membrane protein DDB_G0293934-like isoform X1 | 0.734 |
| RF55_14443 | GB51356 | cytochrome P450 4G11 | 0.726 |
| RF55_5140 | XP_016767978.1 | 0.656 | |
| XLOC_003947 | 0.655 | ||
| XLOC_019296 | -0.643 | ||
| XLOC_005895 | -0.625 | ||
| XLOC_010120 | 0.619 | ||
| RF55_3431 | GB47849 | pyrroline-5-carboxylate reductase 2-like isoform X2 | 0.611 |
| RF55_6542 | GB52074 | 6-phosphogluconate dehydrogenase, decarboxylating | -0.603 |
| RF55_6567 | 552211 | protein THEM6-like | -0.597 |
| RF55_11093 | GB51174 | uncharacterized protein DDB_G0284459-like | 0.593 |
| RF55_9960 | GB52023 | cytochrome P450 6AQ1 isoform X3 | -0.584 |
| RF55_14139 | XP_016768457.1 | 0.561 | |
| RF55_16317 | XP_016770827.1 | -0.523 | |
| RF55_16054 | GB55082 | protein PBDC1-like | 0.489 |
| XLOC_004490 | -0.474 | ||
| RF55_9918 | GB47885 | probable cytochrome P450 304a1 | 0.470 |
| XLOC_015751 | -0.442 | ||
| RF55_12610 | GB40866 | heat shock protein cognate 4 | -0.432 |
| RF55_11067 | GB46772 | very-long-chain enoyl-CoA reductase-like | -0.431 |
| RF55_6451 | GB55598 | troponin I isoform X23 | 0.430 |
| RF55_10641 | XP_016769078.1 | -0.430 | |
| RF55_4656 | GB42792 | uncharacterized protein LOC409805 isoform X3 | 0.415 |
| RF55_4036 | GB44208 | WD repeat-containing protein 37-like isoform X4 | 0.407 |
| XLOC_002901 | -0.398 | ||
| RF55_16927 | XP_006564499.2 | 0.394 | |
| RF55_4554 | GB47990 | tropomyosin-1-like | 0.393 |
| RF55_15079 | GB41028 | ATP synthase subunit alpha, mitochondrial isoform 1 | 0.375 |
| RF55_3507 | GB41333 | DNA-directed RNA polymerase III subunit RPC1-like isoform X1 | -0.375 |
| RF55_5177 | GB43902 | hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like | -0.368 |
| RF55_13988 | GB55263 | putative fatty acyl-CoA reductase CG5065-like | -0.362 |
| RF55_10649 | 0.359 | ||
| RF55_2038 | GB51787 | myosin light chain alkali-like isoform X5 | 0.350 |
| RF55_3707 | GB55537 | transketolase isoform 1 | -0.342 |
| XLOC_005759 | 0.330 | ||
| RF55_10912 | GB47880 | superoxide dismutase 1 | -0.324 |
| RF55_18605 | XP_016770827.1 | -0.315 | |
| RF55_17057 | GB52590 | fatty acid synthase-like isoform 1 | -0.312 |
| RF55_3648 | XP_016768682.1 | -0.310 | |
| RF55_5902 | GB55302 | trehalose transporter 1 isoform X6 | 0.308 |
| RF55_12242 | GB41912 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase-like | -0.286 |
| RF55_10676 | GB40240 | myosin regulatory light chain 2 | 0.279 |
| RF55_4654 | GB45673 | alpha-N-acetylgalactosaminidase-like | 0.277 |
| XLOC_012799 | 0.273 | ||
| XLOC_018477 | -0.268 | ||
| RF55_6754 | XP_016772080.1 | 0.254 | |
| RF55_3967 | GB46039 | tubulin alpha-1 chain-like | -0.251 |
| RF55_1582 | GB45012 | adenosylhomocysteinase-like | 0.248 |
| RF55_2761 | XP_003249233.2 | 0.246 | |
| RF55_2493 | GB45913 | protein lethal(2)essential for life-like | -0.244 |
| RF55_15035 | GB51356 | cytochrome P450 4G11 | -0.223 |
| RF55_5799 | GB50123 | myophilin-like | 0.216 |
| RF55_5341 | GB50508 | fibrillin-2 | -0.215 |
| RF55_598 | GB40021 | probable serine/threonine-protein kinase clkA-like | -0.215 |
| RF55_5109 | XP_016767675.1 | -0.211 | |
| RF55_2407 | XP_016768440.1 | 0.207 | |
| RF55_3343 | GB46290 | acetyl-coenzyme A synthetase-like | -0.205 |
| RF55_10752 | XP_016771487.1 | 0.204 | |
| RF55_19196 | GB41311 | actin, indirect flight muscle-like | 0.202 |
| RF55_5837 | GB43879 | aquaporin AQPcic-like isoform X2 | -0.191 |
| RF55_13604 | XP_016767981.1 | 0.185 | |
| RF55_3994 | GB49175 | 4-hydroxyphenylpyruvate dioxygenase-like | 0.181 |
| RF55_18796 | GB53412 | fatty acid synthase-like | -0.180 |
| XLOC_001770 | 0.175 | ||
| RF55_1934 | GB54827 | synaptotagmin 1 | -0.169 |
| RF55_5219 | GB51753 | uncharacterized protein LOC100576760 isoform X2 | -0.163 |
| RF55_778 | GB42422 | ADP/ATP translocase | 0.154 |
| RF55_10519 | GB54446 | arginine kinase isoform X2 | 0.141 |
| RF55_364 | 0.137 | ||
| RF55_2231 | XP_016773593.1 | -0.134 | |
| XLOC_008839 | -0.132 | ||
| RF55_5431 | -0.129 | ||
| RF55_5198 | XP_016768517.1 | 0.124 | |
| RF55_11370 | XP_016772844.1 | -0.115 | |
| RF55_13251 | GB52590 | fatty acid synthase-like isoform 1 | -0.113 |
| RF55_6842 | GB43052 | paramyosin, long form-like | 0.109 |
| RF55_6077 | GB54423 | uncharacterized protein LOC551958 | -0.108 |
| RF55_6180 | GB40758 | icarapin-like | 0.108 |
| XLOC_005990 | -0.099 | ||
| RF55_10150 | XP_016769706.1 | 0.099 | |
| RF55_1045 | GB54861 | LOW QUALITY PROTEIN: counting factor associated protein D-like | -0.097 |
| RF55_3186 | XP_016770377.1 | 0.092 | |
| XLOC_020265 | -0.089 | ||
| XLOC_019117 | -0.085 | ||
| RF55_4175 | GB44311 | actin related protein 1 | 0.083 |
| XLOC_016272 | -0.079 | ||
| RF55_5206 | GB40735 | fructose-bisphosphate aldolase-like isoform X1 | 0.071 |
| RF55_4024 | XP_016767817.1 | 0.069 | |
| RF55_9453 | GB55096 | NADP-dependent malic enzyme isoform X3 | -0.067 |
| RF55_8355 | GB42809 | translationally-controlled tumor protein homolog isoform 1 | 0.063 |
| RF55_3575 | XP_016768967.1 | -0.062 | |
| XLOC_011290 | 0.061 | ||
| RF55_5559 | GB52107 | tubulin alpha-1 chain-like | 0.061 |
| RF55_3308 | -0.059 | ||
| XLOC_020794 | 0.059 | ||
| XLOC_018966 | 0.059 | ||
| RF55_1104 | XP_016769017.1 | 0.048 | |
| RF55_652 | GB54354 | uncharacterized protein DDB_G0274915-like isoform X2 | 0.045 |
| RF55_3854 | XP_006571125.2 | -0.036 | |
| RF55_6873 | GB47106 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | -0.035 |
| RF55_15158 | GB42794 | circadian clock-controlled protein-like isoform 1 | -0.034 |
| RF55_4888 | GB40779 | transaldolase | 0.034 |
| XLOC_019193 | 0.026 | ||
| XLOC_013126 | -0.025 | ||
| XLOC_017016 | 0.025 | ||
| XLOC_016458 | -0.024 | ||
| XLOC_013131 | -0.013 | ||
| RF55_10886 | GB41427 | catalase | -0.012 |
| XLOC_003165 | -0.010 | ||
| RF55_9333 | GB49688 | peroxidase isoformX2 | 0.008 |
| XLOC_004885 | -0.005 | ||
| RF55_5677 | GB46713 | translation elongation factor 2-like isoform 1 | 0.004 |
| RF55_14822 | GB43823 | chemosensory protein 1 precursor | 0.004 |
| RF55_11002 | XP_016768909.1 | 0.002 |
Genes that are differentially expressed in multiple species
In Tables S6-S7, we have simply listed the genes that showed statistically significant differential expression in two or more species. This method is expected to make few ‘false positive’ errors, but it probably misses many of true overlaps because our study has modest power to detect differential expression (that is, the ‘false negative’ rate is high).
In Table S8, we instead look for similarities between species using a method that ranks genes from most- to least- phermone sensitive based on log fold change, alleviating the problem of low power. To do this, we select the top \(n\) genes per species (where \(n\) = 100, 200… 500), based on the absolute magnitude of the log fold-change response to queen pheromone, giving a list of the most pheromone-sensitive genes. This method produces fewer false negatives in our search for overlapping genes, at cost of increasing the false positive rate.
# Define a function to test whether the overlap of two sets of differentially expressed genes,
# drawn from a common pool (e.g. all the orthologs that were tested), is higher or lower than expected
# Inspiration for this code: https://stats.stackexchange.com/questions/10328/using-rs-phyper-to-get-the-probability-of-list-overlap
overlap.hypergeometric.test <- function(n.overlaps, num.sig1, num.sig2, num.genes, species){
p.smaller <- phyper(n.overlaps, num.sig1, num.genes - num.sig1, num.sig2)
p.higher <- 1 - phyper(n.overlaps - 1, num.sig1, num.genes - num.sig1, num.sig2)
percent_of_maximum_overlaps <- 100 * n.overlaps / min(c(num.sig1, num.sig2))
output <- data.frame(Species = species,
Test = c("Overlap is lower than expected:",
"Overlap is higher than expected:"),
p = c(p.smaller, p.higher),
percent_of_maximum_overlaps = round(percent_of_maximum_overlaps,1))
output[output$p == min(output$p), ]
}
# Apis and flavus
af.oggs <- make.OGGs(c("am", "lf"))[[2]] # Get the orthologous gene list
n.am <- (tbl(my_db, "ebseq_padj_gene_am") %>%
filter(gene %in% af.oggs$am) %>%
summarise(n=n()) %>%
as.data.frame())[1,1] # Count the number of diff expressed genes that appear in the OGG list
n.lf <- (tbl(my_db, "ebseq_padj_gene_lf") %>%
filter(gene %in% af.oggs$lf) %>% summarise(n=n()) %>%
as.data.frame())[1,1]
num.oggs.af <- nrow(af.oggs) # Count the orthologous genes
af.oggs <- suppressMessages(
af.oggs %>%
filter(am %in% (tbl(my_db, "ebseq_padj_gene_am") %>%
as.data.frame())[,1],
lf %in% (tbl(my_db, "ebseq_padj_gene_lf") %>%
as.data.frame())[,1]) %>%
mutate(Species = "Apis and L. flavus") %>%
left_join(tbl(my_db, "ebseq_gene_am") %>%
dplyr::select(gene, PostFC) %>%
rename(am=gene), copy=T) %>%
rename(`Apis FC` = PostFC) %>%
left_join(tbl(my_db, "ebseq_gene_lf") %>%
dplyr::select(gene, PostFC) %>%
rename(lf=gene), copy=T) %>%
rename(`L. flavus FC` = PostFC) %>%
left_join(tbl(my_db, "bee_names") %>%
rename(am=gene), copy=T))
num.overlap.af <- nrow(af.oggs) # Count the overlaps
test1 <- overlap.hypergeometric.test(num.overlap.af,
n.am, n.lf,
num.oggs.af,
"Apis and L. flavus") # Run the hypergeometric test
# Apis and niger
an.oggs <- make.OGGs(c("am", "ln"))[[2]] # Get the orthologous gene list
n.am <- (tbl(my_db, "ebseq_padj_gene_am") %>%
filter(gene %in% an.oggs$am) %>%
summarise(n=n()) %>% as.data.frame())[1,1] # Count the number of diff expressed genes that appear in the OGG list
n.ln <- (tbl(my_db, "ebseq_padj_gene_ln") %>%
filter(gene %in% an.oggs$ln) %>%
summarise(n=n()) %>% as.data.frame())[1,1]
num.oggs.an <- nrow(an.oggs) # Count the orthologous genes
an.oggs <- suppressMessages(
an.oggs %>%
filter(am %in% (tbl(my_db, "ebseq_padj_gene_am") %>% as.data.frame())[,1],
ln %in% (tbl(my_db, "ebseq_padj_gene_ln") %>% as.data.frame())[,1]) %>%
mutate(Species = "Apis and L. niger") %>%
left_join(tbl(my_db, "ebseq_gene_am") %>%
dplyr::select(gene, PostFC) %>%
rename(am=gene), copy=T) %>%
rename(`Apis FC` = PostFC) %>%
left_join(tbl(my_db, "ebseq_gene_ln") %>%
dplyr::select(gene, PostFC) %>%
rename(ln=gene), copy=T) %>%
rename(`L. niger FC` = PostFC) %>%
left_join(tbl(my_db, "bee_names") %>%
rename(am=gene), copy=T))
num.overlap.an <- nrow(an.oggs) # Count the overlaps
test2 <- overlap.hypergeometric.test(num.overlap.an, n.am, n.ln, num.oggs.an, "Apis and L. niger") # Run the hypergeometric test
# flavus and niger
fn.oggs <- make.OGGs(c("lf", "ln"))[[2]] # Get the orthologous gene list
n.lf <- (tbl(my_db, "ebseq_padj_gene_lf") %>%
filter(gene %in% fn.oggs$lf) %>%
summarise(n=n()) %>% as.data.frame())[1,1] # Count the number of diff expressed genes that appear in the OGG list
n.ln <- (tbl(my_db, "ebseq_padj_gene_ln") %>%
filter(gene %in% fn.oggs$ln) %>%
summarise(n=n()) %>% as.data.frame())[1,1]
num.oggs.fn <- nrow(fn.oggs) # Count the orthologous genes
fn.oggs <- suppressMessages(
fn.oggs %>% filter(ln %in% (tbl(my_db, "ebseq_padj_gene_ln") %>% as.data.frame())[,1],
lf %in% (tbl(my_db, "ebseq_padj_gene_lf") %>% as.data.frame())[,1]) %>%
mutate(Species = "L. flavus and L. niger") %>%
left_join(tbl(my_db, "ebseq_gene_lf") %>%
dplyr::select(gene, PostFC) %>%
rename(lf=gene), copy=T) %>%
rename(`L. flavus FC` = PostFC) %>%
left_join(tbl(my_db, "ebseq_gene_ln") %>%
dplyr::select(gene, PostFC) %>%
rename(ln=gene), copy=T) %>%
rename(`L. niger FC` = PostFC) %>%
left_join(tbl(my_db, "lf2am"), copy=T) %>%
left_join(tbl(my_db, "bee_names") %>%
rename(am=gene), copy=T))
num.missing <- sum(is.na(fn.oggs$name))
fn.oggs$name[is.na(fn.oggs$name)] <- paste("Unknown gene", 1:num.missing)
num.overlap.fn <- nrow(fn.oggs) # Count the overlaps
test3 <- overlap.hypergeometric.test(num.overlap.fn,
n.lf, n.ln,
num.oggs.fn,
"L. flavus and L. niger") # Run the hypergeometric test
overlaps <- suppressMessages(
data.frame(name = unique(c(af.oggs$name, an.oggs$name, fn.oggs$name)),
stringsAsFactors = F) %>%
left_join(rbind(af.oggs %>% dplyr::select(name, starts_with("Apis")),
an.oggs %>% dplyr::select(name, starts_with("Apis")))) %>%
left_join(rbind(af.oggs %>% dplyr::select(name, ends_with("flavus FC")),
fn.oggs %>% dplyr::select(name, ends_with("flavus FC")))) %>%
left_join(rbind(an.oggs %>% dplyr::select(name, ends_with("niger FC")),
fn.oggs %>% dplyr::select(name, ends_with("niger FC")))) %>% distinct())
overlaps <- rbind(overlaps[overlaps$name == "myosin light chain alkali-like isoform X5", ],
overlaps[overlaps$name != "myosin light chain alkali-like isoform X5", ])
overlaps$Consistent <- "Yes"
overlaps$Consistent[apply(overlaps[,2:4],1,min,na.rm=T)<1 & apply(overlaps[,2:4],1,max,na.rm=T)>1] <- "No"
for(i in 2:4) overlaps[,i] <- format(round(log2(overlaps[,i]), 3), nsmall = 3)
overlaps[overlaps == " NA"] <- " "
rownames(overlaps) <- NULL
overlap.p.values <- rbind(test1, test2, test3)
rownames(overlap.p.values) <- NULL
all.overlaps <- c(af.oggs$am, an.oggs$am, fn.oggs$am[!is.na(fn.oggs$am)]) %>% uniqueTable S6: All orthologous genes that were significantly differentially expressed between pheromone treatments in more than one species. The FC columns give the Log\(_2\) fold-change in expression for each species where the focal gene was significantly differentially expressed, where positive numbers mean it was expressed at a higher level in control animals. The last column highlights genes that responded to treatment in a consistent or inconsistent direction across species. B. terrestris is omitted because neither of its differentially expressed genes were significantly affected by treatment in the other three species.
saveRDS(overlaps, file = "supplement/tab_S6.rds")
kable.table(overlaps)| name | Apis FC | L. flavus FC | L. niger FC | Consistent |
|---|---|---|---|---|
| myosin light chain alkali-like isoform X5 | 0.444 | 0.372 | 0.350 | Yes |
| proteasome subunit beta type-5-like | 0.484 | -0.434 | No | |
| probable Bax inhibitor 1 | 0.447 | -0.167 | No | |
| intracellular protein transport protein USO1 isoform X2 | -0.684 | 0.445 | No | |
| muscle M-line assembly protein unc-89 isoform X5 | -0.526 | 0.213 | No | |
| transitional endoplasmic reticulum ATPase TER94 | 0.544 | -0.306 | No | |
| actin related protein 1 | 0.762 | 0.083 | Yes | |
| tubulin alpha-1 chain-like | 0.773 | -0.251 | No | |
| Unknown gene 1 | -0.088 | 0.048 | No | |
| uncharacterized protein LOC100576760 isoform X2 | -0.066 | -0.163 | Yes | |
| myosin regulatory light chain 2 | 0.258 | 0.279 | Yes | |
| NADP-dependent malic enzyme isoform X3 | 0.040 | -0.067 | No | |
| transketolase isoform 1 | -0.248 | -0.342 | Yes | |
| troponin T, skeletal muscle | 0.374 | 0.185 | Yes | |
| uncharacterized protein LOC551958 | 0.511 | -0.108 | No | |
| probable cytochrome P450 304a1 | -0.224 | 0.470 | No | |
| Unknown gene 2 | 0.181 | 0.092 | Yes | |
| WD repeat-containing protein 37-like isoform X4 | 0.083 | 0.407 | Yes | |
| Unknown gene 3 | -0.131 | -0.211 | Yes | |
| uncharacterized protein LOC409805 isoform X3 | 0.444 | 0.415 | Yes | |
| ADP/ATP translocase | 0.054 | 0.154 | Yes | |
| peroxidase isoformX2 | 0.104 | 0.008 | Yes | |
| neurofilament heavy polypeptide-like isoform X2 | 0.377 | 0.069 | Yes | |
| fatty acid synthase-like isoform 1 | -0.926 | -1.015 | Yes | |
| LOW QUALITY PROTEIN: counting factor associated protein D-like | -0.294 | -0.097 | Yes | |
| myophilin-like | 0.226 | 0.216 | Yes | |
| Unknown gene 4 | 0.039 | 0.254 | Yes | |
| hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like | 7.514 | -0.368 | No | |
| arginine kinase isoform X2 | 0.441 | 0.141 | Yes | |
| heat shock protein cognate 4 | -0.369 | -0.432 | Yes | |
| tropomyosin-1-like | 0.116 | 0.393 | Yes | |
| 6-phosphogluconate dehydrogenase, decarboxylating | -0.445 | -0.603 | Yes | |
| transaldolase | -0.044 | 0.034 | No | |
| superoxide dismutase 1 | -0.175 | -0.324 | Yes | |
| translation elongation factor 2-like isoform 1 | -0.079 | 0.004 | No | |
| very-long-chain enoyl-CoA reductase-like | -0.201 | -0.431 | Yes | |
| Unknown gene 5 | 0.350 | 0.359 | Yes |
Table S7: The overlap between the lists of significantly differently expressed orthologous genes was significantly higher than expected for L. flavus and L. niger, suggesting that queen pheromone has conserved effects on gene expression between these two species (results based on a hypergeometric test). For the other two species pairs, the number of overlapping genes was not higher or lower than expected under the null hypothesis that queen pheromone affects a random set of genes in each species. The last column gives the number of genes that overlapped, divided by the maximum number that could have overlapped given the numbers of orthologous genes that were significant in each species.
names(overlap.p.values)[4] <- "% of maximum possible overlap"
saveRDS(overlap.p.values, file = "supplement/tab_S7.rds")
pander(overlap.p.values, split.cell = 40, split.table = Inf)| Species | Test | p | % of maximum possible overlap |
|---|---|---|---|
| Apis and L. flavus | Overlap is higher than expected: | 0.1915 | 5.8 |
| Apis and L. niger | Overlap is higher than expected: | 0.2616 | 6.1 |
| L. flavus and L. niger | Overlap is higher than expected: | 0 | 42.3 |
most.pheromone.sensitive.genes <- function(){
ngenes.list <- (1:5)*100 # 100 - 500
overlapping_genes <- list()
permutation_results <- list()
for(i in 1:length(ngenes.list)){
ngenes <- ngenes.list[i]
# Get the top n genes from Apis, as ranked by the absolute value for the log-fold change in response to pheromone
g1 <- tbl(my_db, "ebseq_gene_am") %>% collect() %>%
dplyr::select(gene, PostFC) %>%
mutate(PostFC = log2(PostFC)) %>% left_join(tbl(my_db, "bee_names"), copy=TRUE, by = "gene") %>%
arrange(-abs(PostFC))
# Do the same for genes from non-Apis species, and get the Apis names for the top BLAST hits in the Apis genome
g2 <- tbl(my_db, "ebseq_gene_bt") %>%
dplyr::select(gene, PostFC) %>%
left_join(tbl(my_db, "bt2am") %>%
rename(gene = bt) %>%
dplyr::select(-evalue), by = "gene") %>%
collect() %>% mutate(PostFC = log2(PostFC)) %>%
arrange(-abs(PostFC)) %>%
rename(bt = gene, gene = am) %>%
left_join(tbl(my_db, "bee_names"), copy=TRUE, by = "gene") %>%
filter(!is.na(name))
g3 <- tbl(my_db, "ebseq_gene_lf") %>%
dplyr::select(gene, PostFC) %>%
left_join(tbl(my_db, "lf2am") %>%
rename(gene = lf) %>%
dplyr::select(-evalue), by = "gene") %>%
collect() %>% mutate(PostFC = log2(PostFC)) %>%
arrange(-abs(PostFC)) %>%
rename(lf = gene, gene = am) %>%
left_join(tbl(my_db, "bee_names"), copy=TRUE, by = "gene") %>%
filter(!is.na(name))
g4 <-tbl(my_db, "ebseq_gene_ln") %>%
dplyr::select(gene, PostFC) %>%
left_join(tbl(my_db, "ln2am") %>%
rename(gene = ln) %>%
dplyr::select(-evalue), by = "gene") %>%
collect() %>% mutate(PostFC = log2(PostFC)) %>%
arrange(-abs(PostFC)) %>%
rename(ln = gene, gene = am) %>%
left_join(tbl(my_db, "bee_names"), copy=TRUE, by = "gene") %>%
filter(!is.na(name))
get_top <- function(x, ngenes) {x %>% head(ngenes) %>% .$name %>% unique()}
# Count the number of intersections, and find gene names that appear in 3 or 4 species using the venn() function
venn.diagram <- venn(list(am=(g1 %>% get_top(ngenes)),
bt=(g2 %>% get_top(ngenes)),
lf=(g3 %>% get_top(ngenes)),
ln=(g4 %>% get_top(ngenes))), show.plot=FALSE)
three.plus <- attr(venn.diagram, "intersections")[str_count(names(attr(venn.diagram, "intersections")), ":") > 1] %>%
unlist %>% unname %>% unique %>% sort
four <- attr(venn.diagram, "intersections")[str_count(names(attr(venn.diagram, "intersections")), ":") > 2] %>%
unlist %>% unname %>% unique %>% sort
three <- three.plus[!(three.plus %in% four)]
if(length(three) + length(four) > 0) overlapping_genes[[i]] <- data.frame(Name = c(three, four),
count = c(rep("x3 species", length(three)),
rep("4 species", length(four))),
nGenes = ngenes, stringsAsFactors = FALSE)
count_intersects <- function(l1, l2) {
intersections <- attr(venn(list(am=l1 %>% get_top(ngenes),
bt=l2 %>% get_top(ngenes)),
show.plot=FALSE), "intersections")
if(length(intersections) > 2) return(length(intersections[[3]]))
else return(0)
}
# permutation test
num_overlaps <- c(count_intersects(g1,g2), count_intersects(g1,g3), count_intersects(g1,g4),
count_intersects(g2,g3), count_intersects(g2,g4), count_intersects(g3,g4))
nBoots <- 10000
boot_results <- lapply(1:nBoots, function(x){
g1 <- g1[sample(nrow(g1), nrow(g1)), ]
g2 <- g2[sample(nrow(g2), nrow(g2)), ]
g3 <- g3[sample(nrow(g3), nrow(g3)), ]
g4 <- g4[sample(nrow(g4), nrow(g4)), ]
c(count_intersects(g1,g2), count_intersects(g1,g3), count_intersects(g1,g4),
count_intersects(g2,g3), count_intersects(g2,g4), count_intersects(g3,g4))
}) %>% do.call("rbind", .)
p <- 1:6
for(j in 1:6) p[j] <- sum(boot_results[,j] > num_overlaps[j]) / nBoots
permutation_results[[i]] <- tibble(species_pair = c("am-bt", "am-lf", "am-ln", "bt-lf", "bt-ln", "lf-ln"),
top_n_genes = ngenes.list[i],
observed_overlaps = num_overlaps,
expected_overlaps = colMeans(boot_results),
`O/E` = observed_overlaps / expected_overlaps,
p = p,
x = ifelse(p < 0.05, "sig", " ")) %>% as.data.frame()
} # end of for()
permutation_results <- do.call("rbind", permutation_results)
names(permutation_results) <- c("Species pair", "Size of gene set", "Obs. overlaps", "Exp. overlaps", "O/E", "p-value", "")
overlapping_genes <- do.call("rbind", overlapping_genes) %>%
arrange(count, nGenes, Name) %>%
distinct(paste(Name, count), .keep_all = TRUE) %>%
dplyr::select(Name, count, nGenes) %>%
mutate(count = replace(count, count == "x3 species", "3 species"))
names(overlapping_genes) <- c("Name", "Appears in", "Size of gene set")
list(overlapping_genes, permutation_results)
}
if(!("most.pheromone.sensitive.genes.rds" %in% list.files("data"))){
most.pheromone.sensitive.genes <- most.pheromone.sensitive.genes()
saveRDS(most.pheromone.sensitive.genes, "data/most.pheromone.sensitive.genes.rds")
} else {
most.pheromone.sensitive.genes <- readRDS("data/most.pheromone.sensitive.genes.rds")
}Table S8: List of genes that appear in the top n-most pheromone-sensitive genes for 3 or 4 species. To generate the table, we ranked genes by the absolute value of their log fold change in response to queen pheromone, then listed the gene names that appeared in 3-4 species. For non-Apis species, we found the gene names by comparison with the Apis genome by BLAST. This exercise was performed with n = 100, 200 … 500, and the third column lists the smallest n for which the gene in question appeared (for example, the gene protein takeout-like appeared for all 4 species when inspecting the top 200+ genes).
saveRDS(most.pheromone.sensitive.genes[[1]], file = "supplement/tab_S8.rds")
kable(most.pheromone.sensitive.genes[[1]], "html") %>%
kable_styling() %>%
scroll_box(height = "300px")| Name | Appears in | Size of gene set |
|---|---|---|
| protein takeout-like | 4 species | 200 |
| glucose dehydrogenase [FAD, quinone] | 4 species | 300 |
| histone-lysine N-methyltransferase SETMAR-like | 4 species | 300 |
| serotonin receptor | 4 species | 500 |
| titin-like | 4 species | 500 |
| uncharacterized protein LOC102656088 | 4 species | 500 |
| histone-lysine N-methyltransferase SETMAR-like | 3 species | 200 |
| 2-oxoglutarate dehydrogenase, mitochondrial-like isoform X5 | 3 species | 300 |
| probable serine/threonine-protein kinase DDB_G0282963 isoform X4 | 3 species | 300 |
| titin-like | 3 species | 300 |
| elongation of very long chain fatty acids protein 6-like | 3 species | 400 |
| ligand-gated chloride channel homolog 3 precursor | 3 species | 400 |
| odorant receptor Or2-like | 3 species | 400 |
| putative odorant receptor 13a-like | 3 species | 400 |
| serotonin receptor | 3 species | 400 |
| trypsin-1 | 3 species | 400 |
| uncharacterized protein LOC100576902 | 3 species | 400 |
| uncharacterized protein LOC102655422 | 3 species | 400 |
| uncharacterized protein LOC102656088 | 3 species | 400 |
| metabotropic glutamate receptor 7 isoform X3 | 3 species | 500 |
| probable cytochrome P450 305a1 | 3 species | 500 |
| protein NPC2 homolog | 3 species | 500 |
| suppressor protein SRP40-like | 3 species | 500 |
| uncharacterized protein LOC100577132 | 3 species | 500 |
| uncharacterized protein LOC102656830 | 3 species | 500 |
| uncharacterized protein LOC724216 | 3 species | 500 |
Table S9: Results of a permutation test examining the number of overlaps in the top n-most pheromone-sensitive genes for each pair of species. To generate the table, we ranked genes by the absolute value of their log fold change in response to queen pheromone, then took the top n-most pheromone-sensitive genes for each species, and counted the observed and expected number of overlaps (the expected number was estimated by bootstrapping with 10^5 replicates). The O/E column gives the ratio of observed to expected, where numbers >1 indicate more overlap than expected. The one-tailed p-value was estimated as the proportion of bootstrap replicates showing more overlap than in the real dataset. This exercise was performed with n = 100, 200 … 500.
saveRDS(most.pheromone.sensitive.genes[[2]], file = "supplement/tab_S9.rds")
kable(most.pheromone.sensitive.genes[[2]], "html", digits=2) %>%
kable_styling() %>%
scroll_box(height = "300px")| Species pair | Size of gene set | Obs. overlaps | Exp. overlaps | O/E | p-value | |
|---|---|---|---|---|---|---|
| am-bt | 100 | 1 | 0.97 | 1.03 | 0.25 | |
| am-lf | 100 | 2 | 0.97 | 2.07 | 0.07 | |
| am-ln | 100 | 2 | 0.99 | 2.03 | 0.08 | |
| bt-lf | 100 | 1 | 1.61 | 0.62 | 0.48 | |
| bt-ln | 100 | 4 | 1.65 | 2.42 | 0.02 | sig |
| lf-ln | 100 | 6 | 2.28 | 2.63 | 0.01 | sig |
| am-bt | 200 | 8 | 3.83 | 2.09 | 0.02 | sig |
| am-lf | 200 | 3 | 3.79 | 0.79 | 0.53 | |
| am-ln | 200 | 2 | 3.82 | 0.52 | 0.74 | |
| bt-lf | 200 | 4 | 6.37 | 0.63 | 0.77 | |
| bt-ln | 200 | 14 | 6.59 | 2.13 | 0.00 | sig |
| lf-ln | 200 | 14 | 8.62 | 1.62 | 0.02 | sig |
| am-bt | 300 | 18 | 8.52 | 2.11 | 0.00 | sig |
| am-lf | 300 | 6 | 8.31 | 0.72 | 0.74 | |
| am-ln | 300 | 8 | 8.50 | 0.94 | 0.48 | |
| bt-lf | 300 | 13 | 14.20 | 0.92 | 0.56 | |
| bt-ln | 300 | 28 | 14.67 | 1.91 | 0.00 | sig |
| lf-ln | 300 | 26 | 18.38 | 1.41 | 0.03 | sig |
| am-bt | 400 | 27 | 15.00 | 1.80 | 0.00 | sig |
| am-lf | 400 | 10 | 14.64 | 0.68 | 0.88 | |
| am-ln | 400 | 19 | 14.88 | 1.28 | 0.11 | |
| bt-lf | 400 | 24 | 24.99 | 0.96 | 0.53 | |
| bt-ln | 400 | 47 | 25.74 | 1.83 | 0.00 | sig |
| lf-ln | 400 | 38 | 31.27 | 1.22 | 0.08 | |
| am-bt | 500 | 36 | 23.18 | 1.55 | 0.00 | sig |
| am-lf | 500 | 16 | 22.60 | 0.71 | 0.91 | |
| am-ln | 500 | 22 | 22.90 | 0.96 | 0.52 | |
| bt-lf | 500 | 42 | 38.48 | 1.09 | 0.24 | |
| bt-ln | 500 | 62 | 39.72 | 1.56 | 0.00 | sig |
| lf-ln | 500 | 58 | 47.09 | 1.23 | 0.04 | sig |
Pheromone-sensitive genes tend to be evolutionarily ancient
We defined a gene as “evolutionarily ancient” if it has an ortholog in at least one species from the other lineage (for example, we define ancient A. mellifera genes as those that have a reciprocal best BLAST hit in at least one of the Lasius ant species). Genes that were not detected by BLAST in the other lineage were defined as putatively lineage-specific (though many of them are likely to be conserved genes that we failed to identify as such, hence the word “putative”). Using a non-parametric Mann-Whitney test, we show that the average pheromone sensitivity of ancient genes is significantly lower than that of non-ancient genes, for all 4 species (p < 0.0001). Note that we have probably misclassifed some genes as ancient or lineage-specific, which means that the result might be even stronger than suggested here.
ancient.genes.test <- function(species){
if(species == "am") ancient.genes <- c(make.OGGs(c("am", "lf"))[[2]]$am,
make.OGGs(c("am", "ln"))[[2]]$am) %>% unique
if(species == "bt") ancient.genes <- c(make.OGGs(c("bt", "lf"))[[2]]$bt,
make.OGGs(c("bt", "ln"))[[2]]$bt) %>% unique
if(species == "lf") ancient.genes <- c(make.OGGs(c("lf", "am"))[[2]]$lf,
make.OGGs(c("lf", "bt"))[[2]]$lf) %>% unique
if(species == "ln") ancient.genes <- c(make.OGGs(c("ln", "am"))[[2]]$ln,
make.OGGs(c("ln", "bt"))[[2]]$am) %>% unique
dat <- tbl(my_db, paste("ebseq_gene_", species, sep="")) %>%
collect(n = Inf) %>%
mutate(Ancient = "No",
Ancient = replace(Ancient, gene %in% ancient.genes, "Yes"),
sensitivity = abs(log(PostFC))) %>%
select(Ancient, sensitivity)
ancient.genes <- dat$sensitivity[dat$Ancient == "Yes"]
if(species %in% c("lf", "ln")) {
Putatively.ant.specific.genes <- dat$sensitivity[dat$Ancient == "No"]
print(wilcox.test(ancient.genes, Putatively.ant.specific.genes))
}
else {
Putatively.bee.specific.genes <- dat$sensitivity[dat$Ancient == "No"]
print(wilcox.test(ancient.genes, Putatively.bee.specific.genes))
}
dat %>%
group_by(Ancient) %>%
summarise(Mean_pheromone_sensitivity = mean(sensitivity), SE = sd(sensitivity) / sqrt(length(sensitivity))) %>% pander()
}Apis mellifera
ancient.genes.test("am")
Wilcoxon rank sum test with continuity correction
data: ancient.genes and Putatively.bee.specific.genes
W = 13256000, p-value < 2.2e-16
alternative hypothesis: true location shift is not equal to 0| Ancient | Mean_pheromone_sensitivity | SE |
|---|---|---|
| No | 0.39 | 0.004841 |
| Yes | 0.2368 | 0.002991 |
Bombus terrestris
ancient.genes.test("bt")
Wilcoxon rank sum test with continuity correction
data: ancient.genes and Putatively.bee.specific.genes
W = 8329700, p-value < 2.2e-16
alternative hypothesis: true location shift is not equal to 0| Ancient | Mean_pheromone_sensitivity | SE |
|---|---|---|
| No | 0.2277 | 0.005443 |
| Yes | 0.1084 | 0.001806 |
Lasius flavus
ancient.genes.test("lf")
Wilcoxon rank sum test with continuity correction
data: ancient.genes and Putatively.ant.specific.genes
W = 74676000, p-value < 2.2e-16
alternative hypothesis: true location shift is not equal to 0| Ancient | Mean_pheromone_sensitivity | SE |
|---|---|---|
| No | 0.316 | 0.001468 |
| Yes | 0.1269 | 0.002151 |
Lasius niger
ancient.genes.test("ln")
Wilcoxon rank sum test with continuity correction
data: ancient.genes and Putatively.ant.specific.genes
W = 23871000, p-value < 2.2e-16
alternative hypothesis: true location shift is not equal to 0| Ancient | Mean_pheromone_sensitivity | SE |
|---|---|---|
| No | 0.1761 | 0.00145 |
| Yes | 0.1004 | 0.001726 |
Correlations in pheromone-sensitive gene expression across species
These tests ask if pairs of orthologous genes tend to show a similar level of pheromone sensitivity across species. These tests ask whether there is a cross-species correlation in absolute log fold change in response to pheromone treatment, as calculated using ebseq. The advantage of these tests is that one is not limited to focusing on the set of ‘significant’ genes (as with the Venn diagrams shown in Figure 1), which discards a lot of information becuase it uses an arbitrary p-value threshold, and puts all the genes into two bins (ignoring the quantitative differences in fold-change).
Define function to retrieve fold-change expression data (calculated with ebseq) for genes that are each other’s reciprocal best BLAST
get.fc.for.pair.species <- function(species1, species2){
query <- 'SELECT rbb.X AS X_gene, rbb.Y AS Y_gene, ebseq_gene_X.PostFC AS X_fc, ebseq_gene_Y.PostFC AS Y_fc FROM ebseq_gene_X
JOIN
(SELECT Y2X.X, Y2X.Y FROM Y2X
JOIN X2Y
ON Y2X.Y = X2Y.Y AND Y2X.X = X2Y.X ) AS rbb
ON rbb.X = ebseq_gene_X.gene
JOIN ebseq_gene_Y
ON rbb.Y = ebseq_gene_Y.gene'
query <- str_replace_all(query, "X", species1)
query <- str_replace_all(query, "Y", species2)
dbGetQuery(db, query)
}Make a table and some plots of the correlations for each pair of species
Table S10: Results of Spearman’s rank correlations, testing whether the absolute log fold difference between pheromones treatments is correlated for a given pair of species. Positive coefficients (\(\rho\), written as rho) indicate that on average, orthologous genes have similar sensitivity to queen pheromones. The p-values have been corrected for multiple testing with the Benjamini-Hochberg method.
species.combinations <- t(combn(c("am", "bt", "lf", "ln"), 2))
rho <- numeric(nrow(species.combinations))
p <- numeric(nrow(species.combinations))
for(i in 1:nrow(species.combinations)){
fc.data <- get.fc.for.pair.species(species.combinations[i, 1],
species.combinations[i, 2])
fc.data[,3:4] <- fc.data[,3:4] %>% log2 %>% abs # Log the fold changes - not that it matters for Spearman's!
results <- suppressWarnings(with(fc.data, cor.test(fc.data[,3],
fc.data[,4], method="spearman"))) # Spearman's correlation
rho[i] <- results$estimate
p[i] <- results$p.value
}
species.combinations <- data.frame(Species1 = species.combinations[,1],
Species2 = species.combinations[,2],
rho = rho,
p = p.adjust(p, method = "BH"), # adjust these p-vals for multiple testing
sig = " ", stringsAsFactors = F)
species.combinations[species.combinations == "am"] <- "Apis mellifera"
species.combinations[species.combinations == "bt"] <- "Bombus terrestris"
species.combinations[species.combinations == "lf"] <- "Lasius flavus"
species.combinations[species.combinations == "ln"] <- "Lasius niger"
species.combinations$sig[species.combinations$p < 0.05] <- "*"
species.combinations$sig[species.combinations$p < 0.01] <- "**"
species.combinations$sig[species.combinations$p < 0.0001] <- "***"
# Make a table
saveRDS(species.combinations %>% mutate(rho = format(round(rho, 3), nsmall = 3)), file = "supplement/tab_S10.rds")
species.combinations %>% mutate(rho = format(round(rho, 3), nsmall = 3)) %>% pander| Species1 | Species2 | rho | p | sig |
|---|---|---|---|---|
| Apis mellifera | Bombus terrestris | 0.136 | 3.232e-24 | *** |
| Apis mellifera | Lasius flavus | 0.100 | 1.478e-12 | *** |
| Apis mellifera | Lasius niger | 0.077 | 1.853e-07 | *** |
| Bombus terrestris | Lasius flavus | 0.159 | 3.112e-39 | *** |
| Bombus terrestris | Lasius niger | 0.127 | 2.282e-24 | *** |
| Lasius flavus | Lasius niger | 0.194 | 3.765e-61 | *** |
# Make a heat map showing the correlations
heat.map.figure <- species.combinations %>% ggplot(aes(Species1, Species2, fill=rho)) +
geom_tile(colour="white", size = 4) +
scale_fill_gradient2(name = "Corr",
low = brewer.pal(9, "RdYlBu")[7],
mid = "white",
high = brewer.pal(9, "RdYlBu")[2]) +
geom_text(aes(label = paste(format(signif(rho, 2), nsmall = 2), sig)), size = 3.5) +
theme_bw() + theme(panel.grid = element_blank(),
panel.border = element_blank(),
axis.ticks = element_blank(),
axis.text = element_text(face = "italic"),
legend.position = "none") +
xlab(NULL) + ylab(NULL) +
scale_x_discrete(labels = c("Apis\nmellifera", "Bombus\nterrestris", "Lasius\nflavus"), expand = c(0,0)) +
scale_y_discrete(labels = c("Bombus\nterrestris", "Lasius\nflavus", "Lasius\nniger"), expand = c(0,0))
heat.map.figure
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
ggsave(heat.map.figure, file = "figures/Correlations heat map.pdf", height = 4, width=4.3)# Make a scatterplot showing the correlations
combos <- t(combn(c("am", "bt", "lf", "ln"), 2))
scatter_plot_data <- list()
for(i in 1:nrow(combos)){
fc.data <- get.fc.for.pair.species(combos[i, 1],
combos[i, 2])
fc.data <- fc.data[,3:4] %>% log2 %>% abs # Log the fold changes
names(fc.data) <- c("x", "y")
scatter_plot_data[[i]] <- data.frame(fc.data, sp1 = species.combinations[i, 1], sp2 = species.combinations[i, 2])
}
fig_S3 <- scatter_plot_data %>%
do.call("rbind", .) %>%
ggplot(aes(x, y)) +
geom_point(alpha = 0.15) +
stat_smooth(method = "lm", colour = "tomato") +
facet_wrap(~paste(sp1, sp2, sep = " v "), scales = "free", ncol = 2) +
labs(x = "Pheromone sensitivity of ortholog in species 1",
y = "Pheromone sensitivity of ortholog in species 2") +
theme_minimal()
saveRDS(fig_S3, file = "supplement/fig_S3.rds")
fig_S3
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Figure S3: Each scatterplot shows the correlation in pheromone sensitivity across pairs of orthologous genes, for each of the six possible species pairs. Species 1 refers to the first-listed species, such that in the top-left panel, Apis mellifera is plotted on the x-axis and Bombus terrestris is on the y-axis. The regression lines are from a simple linear regression, and the grey zone around the line shows its 95% confidence intervals.
Identifying genes that splice differently following queen pheromone treatment
Many genes showed alternative splicing in all four species, with the great majority showing between 1 and 6 isoforms.
fig_S4 <- rbind(my_db %>% tbl("isoforms_am") %>% collect %>% .$gene %>%
table %>% melt %>% dplyr::select(value) %>% mutate(sp = "Apis mellifera"),
my_db %>% tbl("isoforms_bt") %>% collect %>% .$gene %>%
table %>% melt %>% dplyr::select(value) %>% mutate(sp = "Bombus terrestris"),
my_db %>% tbl("isoforms_lf") %>% collect %>% .$gene %>%
table %>% melt %>% dplyr::select(value) %>% mutate(sp = "Lasius niger"),
my_db %>% tbl("isoforms_ln") %>% collect %>% .$gene %>%
table %>% melt %>% dplyr::select(value) %>% mutate(sp = "Lasius flavus")) %>%
as.data.frame() %>%
ggplot(aes(x = value, y = sp, fill = sp)) +
geom_joy(stat = "binline", binwidth=1) +
coord_cartesian(xlim=c(1,12)) +
scale_fill_cyclical(values = c("#4040B0", "#9090F0")) +
scale_x_continuous(breaks = seq(1,12,by=1)) +
scale_y_discrete(expand = c(0.01, 0)) +
theme_joy() +
xlab("Number of isoforms") + ylab(NULL) +
theme(axis.text = element_text(face = "italic"))
saveRDS(fig_S4, file = "supplement/fig_S4.rds")
fig_S4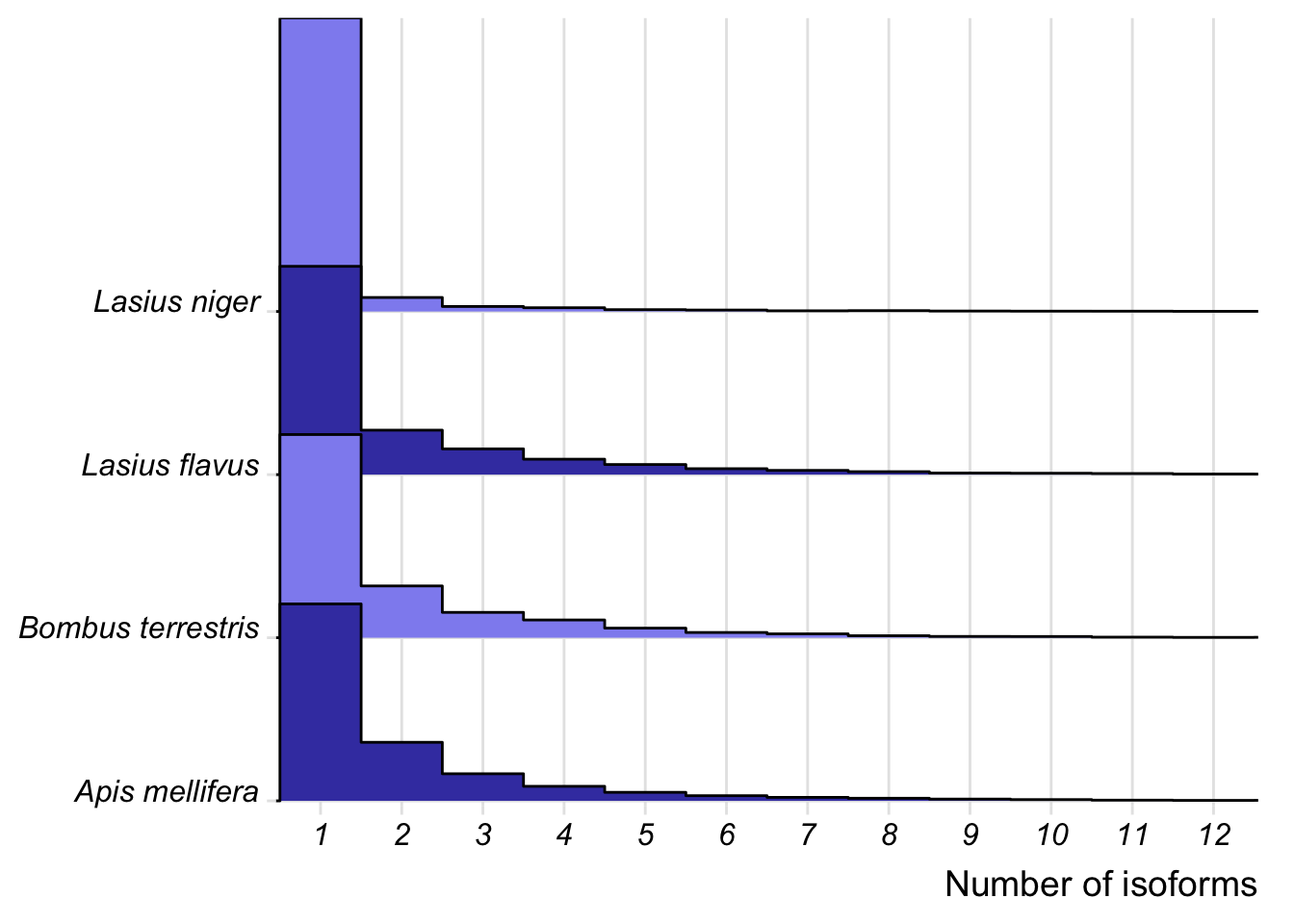
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Figure S4: Distribution of isoform numbers per gene for each of the four species.
# Define a function to search for significantly alternatively spliced genes. We define these as genes that have at least two differentially expressed isoforms, where queen pheromone stimulates expresison of one isoform but surpresses expression of another
find.alternatively.spliced.genes <- function(species){
# Write a database query with 'X' as a placeholder for the species name
# We want to find genes in which at least two isoforms are differentially expressed in response to pheromones
# Significant isoforms are listed in the ebseq_padj_isoform_X tables, so we count them and get the names of their genes
query <- 'SELECT isoforms_X.gene, isoforms_X.isoform, PostFC FROM isoforms_X
JOIN
(SELECT gene, COUNT(*) FROM ebseq_padj_isoform_X
JOIN isoforms_X
ON isoforms_X.isoform = ebseq_padj_isoform_X.isoform
GROUP BY gene
HAVING COUNT(*)>1 ) AS multiISO
ON multiIso.gene = isoforms_X.gene
JOIN ebseq_padj_isoform_X
ON ebseq_padj_isoform_X.isoform = isoforms_X.isoform
ORDER BY isoforms_X.gene'
query <- str_replace_all(query, "X", species) # replace X with the species
Splice <- dbGetQuery(db, query) # run the query
# Find the highest and lowest fold change for the isoforms of each gene
SpliceMin <- aggregate(Splice$PostFC, by=list(Splice$gene), FUN="min")
SpliceMax <- aggregate(Splice$PostFC, by=list(Splice$gene), FUN="max")
SpliceCompare <- merge(SpliceMin, SpliceMax, by = "Group.1")
colnames(SpliceCompare) <- c("gene", "min", "max")
# Define diff-spliced genes as those with both elevated and
# repressed isoforms, when comparing control and pheromone-treated workers
output <- data.frame(gene = SpliceCompare$gene[SpliceCompare$min < 1 & SpliceCompare$max > 1], stringsAsFactors = F)
# Add the Apis mellifera ortholog, from one-way BLAST, if it's a non-Apis species
if(species != "am"){
orthologs <- tbl(my_db, paste(species, "2am", sep = "")) %>% select(-evalue) %>% collect(n=Inf) %>% as.data.frame()
names(orthologs) <- c("gene", "Amel_ortholog")
output <- left_join(output, orthologs, by = "gene")
output <- left_join(output, tbl(my_db, "bee_names") %>%
rename(Amel_ortholog = gene), copy=T, by = "Amel_ortholog")
}
else output <- left_join(output, tbl(my_db, "bee_names"), by = "gene", copy = T)
output[is.na(output)] <- " "
# Remove the 'isoform X' part from the gene name, so it gives the name of the gene not the isoform
output$name <- unlist(lapply(strsplit(output$name, split = " isoform "), function(x) x[1]))
output <- left_join(output, SpliceCompare, by = "gene")
output$min <- log2(output$min) # Log2 transform the fold changes
output$max <- log2(output$max)
output <- output %>% arrange(-(abs(min) + max)) # Arrange by effect size
if(species == "am" | species == "bt") names(output) <- c("Gene", "Name", "Lowest FC", "Highest FC")
else names(output) <- c("Gene", "Amel ortholog", "Name", "Lowest FC", "Highest FC")
output
}Genes showing significant pheromone-induced alternative splicing
There were no genes showing significant pheromone-sensitive splicing for B. terrestris, hence this section contains only three tabs.
A. mellifera
Table S11: List of genes showing statistically significant pheromone-induced alternative splicing in A. mellifera. These genes were defined as those that have at least two isoforms that are differentially expressed following pheromone treatment (EBseq; posterior probability of differential expression p < 0.05), and for which one isoform increases in expression while another decreases. The last two columns show the fold changes of the most down-regulated and most up-regulated isoforms, on a Log\(_2\) scale.
am.alt.splice <- find.alternatively.spliced.genes("am")
saveRDS(am.alt.splice, file = "supplement/tab_S11.rds")
kable.table(am.alt.splice)| Gene | Name | Lowest FC | Highest FC |
|---|---|---|---|
| GB44254 | uncharacterized protein LOC411586 | -10.2106382 | 7.3294705 |
| GB55598 | troponin I | -8.6184698 | 7.4257311 |
| GB55650 | ryanodine receptor 44F | -8.6521541 | 6.1211069 |
| GB42000 | cAMP-specific 3’,5’-cyclic phosphodiesterase, isoforms N/G | -7.8461925 | 6.4652082 |
| GB49567 | sestrin-1-like isoformX1 | -6.7384430 | 6.3303655 |
| GB46113 | uncharacterized protein LOC726188 | -6.7215768 | 6.2049532 |
| GB55237 | disco-interacting protein 2 | -6.4126080 | 5.7870758 |
| GB44556 | uncharacterized protein LOC411962 | -5.3193508 | 6.4264182 |
| GB50923 | serine-protein kinase ATM | -5.4981139 | 6.1549064 |
| GB50941 | phosphatidate phosphatase LPIN2-like | -6.1378916 | 5.3694436 |
| GB55848 | DNA ligase 1-like | -6.0254223 | 5.3645877 |
| GB42142 | nuclear hormone receptor FTZ-F1 | -6.0475859 | 5.2543590 |
| GB45259 | zinc finger protein 91-like | -5.7141149 | 5.4922565 |
| GB52595 | zinc finger and BTB domain-containing protein 20-like | -6.0058327 | 5.1479879 |
| GB55225 | sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 | -3.5827048 | 7.3608415 |
| GB55102 | F-box only protein 28-like | -5.6780118 | 5.1328776 |
| GB42895 | trichohyalin-like | -6.0749616 | 4.6900449 |
| GB44373 | protein zyg-11 homolog B-like | -2.7153166 | 7.8600431 |
| GB49111 | neuropathy target esterase sws | -1.8617078 | 8.3596119 |
| GB46271 | protein BCL9 homolog | -3.1471390 | 6.9356291 |
| GB53431 | inositol hexakisphosphate kinase 2-like | -6.3417004 | 3.3365701 |
| GB44336 | protein couch potato-like | -2.1830854 | 7.3541749 |
| GB45277 | multidrug resistance-associated protein 4-like | -8.1135336 | 1.3365168 |
| GB40263 | dentin sialophosphoprotein-like | -3.0597817 | 6.2723878 |
| 725417 | uncharacterized protein LOC725417 | -2.8308639 | 6.1603441 |
| GB55517 | uncharacterized protein LOC410000 | -3.2763532 | 5.3768010 |
| GB55429 | CREB-regulated transcription coactivator 1-like | -5.3728279 | 3.1844409 |
| GB55998 | beta-1,4-N-acetylgalactosaminyltransferase bre-4 | -2.7284140 | 5.5956924 |
| GB52604 | LIM domain-binding protein 2-like | -2.2872276 | 5.8679044 |
| GB41734 | reversion-inducing-cysteine-rich protein with kazal motifs | -5.9697261 | 1.9393374 |
| GB49535 | calcium/calmodulin-dependent protein kinase II | -1.9949124 | 5.8059893 |
| GB45662 | conserved oligomeric Golgi complex subunit 4-like | -5.3583810 | 2.1410794 |
| GB40310 | uncharacterized protein LOC411575 | -0.9447237 | 6.4575407 |
| GB51117 | rho GTPase-activating protein 18-like | -0.9711563 | 6.3563951 |
| GB41129 | protein LMBR1L-like | -5.6199019 | 1.4425609 |
| GB44876 | uncharacterized protein LOC100576421 | -5.3652303 | 1.6410521 |
| GB50244 | NHL repeat-containing protein 2 | -0.9371409 | 5.9580141 |
| GB47138 | calcium-activated potassium channel slowpoke-like | -0.9035332 | 5.9504672 |
| GB50099 | uncharacterized abhydrolase domain-containing protein DDB_G0269086-like | -1.5085077 | 5.0811380 |
| GB41908 | PERQ amino acid-rich with GYF domain-containing protein CG11148-like | -1.0050181 | 5.5164280 |
| GB52999 | neurofilament heavy polypeptide-like | -2.7388052 | 3.6790149 |
| GB51651 | putative inorganic phosphate cotransporter-like | -5.4170376 | 0.9305746 |
| GB50877 | tyrosine-protein kinase Dnt | -2.5061716 | 3.1568560 |
| GB43220 | transcription termination factor 2 | -3.2229375 | 1.8337147 |
| GB40565 | ribosomal protein S6 kinase alpha-5 | -2.3132702 | 2.6623092 |
| GB55467 | neural cell adhesion molecule L1-like | -3.3341247 | 1.4565432 |
| GB48034 | protein numb-like, transcript variant X5 | -1.0127583 | 3.7699557 |
| GB55570 | transmembrane protein 53-like | -2.0692979 | 2.6556081 |
| GB49417 | PITH domain-containing protein GA19395-like | -2.1836294 | 2.2457448 |
| GB43282 | guanine nucleotide-binding protein G(q) subunit alpha-like | -2.3600303 | 2.0113196 |
| GB48573 | probable multidrug resistance-associated protein lethal(2)03659-like | -2.1285308 | 1.9867141 |
| GB50366 | uncharacterized protein LOC551450 | -0.8408050 | 1.1435481 |
| GB46121 | ubiquitin fusion degradation protein 1 homolog | -0.9511837 | 0.9182434 |
| GB52052 | venom carboxylesterase-6-like | -0.7021931 | 0.9102424 |
L. flavus
Table S12: List of genes showing statistically significant pheromone-induced alternative splicing in Lasius flavus. These genes were defined as those that have at least two isoforms that are differentially expressed following pheromone treatment (EBseq; posterior probability of differential expression p < 0.05), and for which one isoform increases in expression while another decreases. The last two columns show the fold changes of the most down-regulated and most up-regulated isoforms, on a Log\(_2\) scale.
lf.alt.splice<- find.alternatively.spliced.genes("lf")
saveRDS(lf.alt.splice, file = "supplement/tab_S12.rds")
kable.table(lf.alt.splice)| Gene | Amel ortholog | Name | Lowest FC | Highest FC |
|---|---|---|---|---|
| TRINITY_DN11346_c0_g1 | GB49250 | heme oxygenase | -9.1733822 | 4.2807355 |
| TRINITY_DN14030_c0_g1 | XP_016769715.1 | -4.5698062 | 6.1058670 | |
| TRINITY_DN13728_c1_g1 | GB44606 | AMP deaminase 2-like | -5.6147192 | 4.2278104 |
| TRINITY_DN13759_c0_g1 | XP_016766389.1 | -3.7805082 | 5.5650278 | |
| TRINITY_DN13897_c0_g1 | GB41293 | histone acetyltransferase KAT8 | -4.6420034 | 2.6858828 |
| TRINITY_DN14126_c1_g2 | GB43039 | restin homolog | -1.8789713 | 5.3598354 |
| TRINITY_DN12581_c0_g2 | XP_016767063.1 | -4.1075789 | 2.7973408 | |
| TRINITY_DN14084_c0_g1 | XP_016772396.1 | -2.4056262 | 4.1607373 | |
| TRINITY_DN13929_c0_g1 | GB53852 | paired amphipathic helix protein Sin3a | -2.0878837 | 4.4200505 |
| TRINITY_DN13956_c0_g1 | GB54341 | RNA-binding protein 33-like | -1.9836823 | 4.5150571 |
| TRINITY_DN13699_c0_g1 | GB41835 | sorting nexin-13-like | -2.5195188 | 3.9141700 |
| TRINITY_DN13676_c0_g1 | XP_016767354.1 | -3.9853775 | 2.2791646 | |
| TRINITY_DN13889_c0_g1 | GB44338 | small G protein signaling modulator 3 homolog | -2.6671307 | 3.5553713 |
| TRINITY_DN11861_c1_g1 | GB43504 | neural/ectodermal development factor IMP-L2 | -2.2204311 | 3.4241368 |
| TRINITY_DN14001_c0_g2 | GB41033 | facilitated trehalose transporter Tret1-like | -3.4253663 | 2.1957523 |
| TRINITY_DN12063_c0_g1 | GB49936 | calcium uptake protein 1 homolog, mitochondrial-like | -1.9568877 | 3.6347798 |
| TRINITY_DN13321_c0_g1 | XP_016769905.1 | -2.8397819 | 2.7402835 | |
| TRINITY_DN13931_c4_g1 | GB47270 | cytochrome P450 4C1 | -2.2502914 | 2.8866039 |
| TRINITY_DN12721_c1_g1 | GB43391 | transmembrane protein 8B-like | -2.1793408 | 2.8228945 |
| TRINITY_DN11401_c0_g1 | GB50510 | uncharacterized protein LOC409674 | -2.6928098 | 1.9864175 |
| TRINITY_DN10695_c0_g1 | XP_016773356.1 | -2.1193002 | 2.4882173 | |
| TRINITY_DN13698_c3_g1 | GB45025 | mTERF domain-containing protein 1, mitochondrial-like | -1.9661074 | 2.5552928 |
| TRINITY_DN13919_c0_g1 | GB49593 | cytosolic carboxypeptidase-like protein 5-like | -1.6893918 | 2.7745341 |
| TRINITY_DN14156_c10_g1 | XP_016768402.1 | -2.0840677 | 2.3753245 | |
| TRINITY_DN13974_c0_g1 | XP_016768210.1 | -1.5945919 | 2.7670315 | |
| TRINITY_DN12596_c0_g1 | GB40801 | thioredoxin-like protein 4A-like | -1.7080464 | 2.3372385 |
| TRINITY_DN13248_c0_g1 | GB42981 | beta-1,3-glucan-binding protein | -1.6951844 | 2.3160927 |
| TRINITY_DN14247_c8_g2 | GB52590 | fatty acid synthase-like | -1.0570339 | 1.0113553 |
| TRINITY_DN10832_c0_g1 | GB54056 | serine hydroxymethyltransferase, cytosolic | -1.2534345 | 0.3609471 |
| TRINITY_DN11786_c1_g1 | GB51214 | troponin T, skeletal muscle | -0.4087698 | 0.8466319 |
| TRINITY_DN13339_c0_g1 | GB40735 | fructose-bisphosphate aldolase-like | -0.1714515 | 0.3993615 |
| TRINITY_DN13721_c8_g1 | XP_016772716.1 | -0.2069941 | 0.2124566 | |
| TRINITY_DN13271_c0_g1 | GB40312 | choline/ethanolamine kinase-like | -0.1549773 | 0.0102486 |
| TRINITY_DN10322_c0_g1 | XP_016769014.1 | -0.1131430 | 0.0289235 |
L. niger
Table S13: List of genes showing statistically significant pheromone-induced alternative splicing in Lasius flavus. These genes were defined as those that have at least two isoforms that are differentially expressed following pheromone treatment (EBseq; posterior probability of differential expression p < 0.05), and for which one isoform increases in expression while another decreases. The last two columns show the fold changes of the most down-regulated and most up-regulated isoforms, on a Log\(_2\) scale.
ln.alt.splice <- find.alternatively.spliced.genes("ln")
saveRDS(ln.alt.splice, file = "supplement/tab_S13.rds")
kable.table(ln.alt.splice)| Gene | Amel ortholog | Name | Lowest FC | Highest FC |
|---|---|---|---|---|
| RF55_752 | GB48208 | protein argonaute-2 | -3.2926101 | 6.4472781 |
| RF55_883 | XP_016770988.1 | -5.9073798 | 3.5987463 | |
| RF55_4195 | XP_016772612.1 | -5.1744722 | 4.3092261 | |
| RF55_412 | XP_016770021.1 | -3.3875926 | 5.9670345 | |
| RF55_2523 | XP_016770619.1 | -4.7649090 | 4.0350580 | |
| RF55_3907 | XP_016771611.1 | -4.4508904 | 3.6861314 | |
| RF55_4516 | GB54906 | niemann-Pick C1 protein-like | -3.7980002 | 4.3024830 |
| RF55_2336 | GB54910 | tyrosine-protein kinase Abl-like | -3.2249951 | 4.4534584 |
| RF55_1192 | XP_016769850.1 | -1.9480198 | 5.7297004 | |
| XLOC_016164 | -5.4972688 | 2.1282740 | ||
| RF55_3254 | XP_016773300.1 | -2.7312184 | 4.8910736 | |
| RF55_1559 | XP_016768510.1 | -2.9908389 | 4.3225630 | |
| RF55_4335 | XP_016767803.1 | -3.8099604 | 3.4311823 | |
| RF55_6516 | GB40718 | thioredoxin reductase 1 | -3.2537489 | 3.9222290 |
| RF55_11163 | GB53163 | transient receptor potential channel pyrexia | -3.9015904 | 3.2568624 |
| RF55_1926 | XP_016766364.1 | -1.7898846 | 5.3146852 | |
| RF55_9605 | GB42484 | aminopeptidase N-like isoformX1 | -5.0263823 | 2.0370079 |
| XLOC_003165 | -0.6701274 | 6.3901363 | ||
| RF55_4472 | GB43953 | cycle | -3.9049333 | 3.0344073 |
| RF55_13096 | XP_016767210.1 | -4.8346835 | 2.0905931 | |
| RF55_9014 | GB55507 | FYVE, RhoGEF and PH domain-containing protein 4-like | -3.9205477 | 2.9639182 |
| RF55_8972 | XP_016771571.1 | -4.8894254 | 1.9830903 | |
| RF55_4341 | XP_392463.4 | -3.2324663 | 3.6096634 | |
| XLOC_013280 | -2.4206648 | 4.3918399 | ||
| RF55_2163 | GB55475 | cullin-5 | -1.9499220 | 4.8473456 |
| RF55_1574 | XP_016770060.1 | -0.5327703 | 6.2375585 | |
| RF55_6406 | XP_016767146.1 | -2.0553009 | 4.6650121 | |
| RF55_1819 | GB51068 | arrestin homolog | -2.3004627 | 4.3994752 |
| RF55_2357 | XP_016769793.1 | -4.9713497 | 1.7125669 | |
| RF55_2452 | XP_016767413.1 | -4.9414495 | 1.6803943 | |
| RF55_764 | XP_001121384.4 | -4.4683077 | 2.1308271 | |
| RF55_5343 | XP_016766531.1 | -3.3768061 | 3.2009350 | |
| RF55_2401 | XP_016769193.1 | -3.8965864 | 2.6724345 | |
| RF55_6625 | XP_016770656.1 | -2.0711259 | 4.4766181 | |
| RF55_4021 | XP_016767419.1 | -1.8167853 | 4.6555595 | |
| RF55_317 | XP_016769975.1 | -2.7140799 | 3.7123571 | |
| RF55_561 | GB51542 | PH-interacting protein | -3.3596059 | 2.9952497 |
| RF55_2755 | GB48836 | uncharacterized protein LOC100577578 | -4.5392256 | 1.7981131 |
| RF55_357 | -4.0137355 | 2.2850079 | ||
| RF55_9274 | XP_016767607.1 | -4.1097469 | 2.1003576 | |
| RF55_1902 | GB46211 | zinc finger protein 665-like | -4.2211377 | 1.9522150 |
| RF55_1336 | XP_016767701.1 | -4.2155289 | 1.9317851 | |
| RF55_1520 | XP_016768717.1 | -2.0486448 | 4.0217111 | |
| RF55_2217 | GB47260 | zinc finger protein 598-like | -1.9608779 | 4.1054197 |
| RF55_6360 | XP_003251881.3 | -3.5776193 | 2.4733234 | |
| RF55_15946 | XP_016767569.1 | -2.3204379 | 3.7235457 | |
| RF55_4138 | XP_016766515.1 | -2.3589464 | 3.6642057 | |
| RF55_1328 | GB41746 | neogenin | -2.0301217 | 3.8766286 |
| RF55_6748 | GB52716 | NAD kinase-like, transcript variant X11 | -3.4936440 | 2.3932969 |
| RF55_7315 | GB55434 | rab GDP dissociation inhibitor beta | -3.1288936 | 2.7150106 |
| RF55_9900 | XP_016767864.1 | -3.5513206 | 2.2796009 | |
| RF55_1325 | GB44695 | lysophospholipase-like protein 1-like | -4.0976819 | 1.6743676 |
| RF55_9036 | XP_016768958.1 | -3.9412444 | 1.7818052 | |
| RF55_742 | XP_016769412.1 | -2.7605014 | 2.9581890 | |
| RF55_9907 | GB47039 | dynamin | -3.9896574 | 1.7131009 |
| RF55_9185 | XP_016768847.1 | -3.8364223 | 1.8598316 | |
| RF55_2976 | GB42054 | sodium/potassium-transporting ATPase subunit alpha | -2.0568520 | 3.6039514 |
| RF55_4649 | GB43618 | aconitate hydratase, mitochondrial-like | -5.0413696 | 0.5716779 |
| RF55_12355 | GB42664 | probable ATP-dependent RNA helicase DDX43-like | -1.9943294 | 3.6184641 |
| RF55_1934 | GB54827 | synaptotagmin 1 | -5.3908347 | 0.1905938 |
| RF55_360 | GB46768 | uncharacterized MFS-type transporter C09D4.1-like | -3.2312572 | 2.3204411 |
| RF55_5123 | XP_016772933.1 | -2.7363919 | 2.8109308 | |
| RF55_3009 | GB53317 | dentin sialophosphoprotein-like | -3.7638023 | 1.7787767 |
| RF55_5073 | GB55540 | zinc finger MYM-type protein 3-like | -1.8616556 | 3.6633963 |
| RF55_9319 | XP_016768793.1 | -1.6023217 | 3.8961635 | |
| RF55_952 | GB42014 | E3 ubiquitin-protein ligase TRIP12-like | -1.6637152 | 3.6314244 |
| RF55_3561 | GB41659 | endothelin-converting enzyme 1 | -1.9116224 | 3.3339080 |
| RF55_4175 | GB44311 | actin related protein 1 | -1.4407595 | 3.7728266 |
| RF55_7804 | -2.5441138 | 2.6300085 | ||
| RF55_1689 | XP_016768411.1 | -3.3768014 | 1.7465719 | |
| RF55_5052 | XP_016773337.1 | -2.4182694 | 2.6840874 | |
| RF55_457 | GB40931 | uncharacterized protein LOC409781 | -2.3507186 | 2.6953986 |
| RF55_7632 | XP_016767095.1 | -2.0462508 | 2.9941556 | |
| RF55_11518 | GB41804 | nardilysin | -2.2810020 | 2.7512083 |
| RF55_4355 | XP_016771542.1 | -2.4450723 | 2.5183903 | |
| RF55_1518 | XP_016768753.1 | -3.0929627 | 1.8614463 | |
| RF55_5804 | XP_016766957.1 | -2.6227377 | 2.2732244 | |
| RF55_4425 | GB51264 | glutamine-dependent NAD(+) synthetase, transcript variant X3 | -2.1938416 | 2.6792288 |
| RF55_5947 | XP_016768497.1 | -2.0359071 | 2.6987690 | |
| RF55_3209 | GB46684 | monocarboxylate transporter 3-like | -2.2501560 | 2.4459333 |
| RF55_4132 | XP_006572145.2 | -1.8924997 | 2.7949094 | |
| RF55_3822 | XP_016769466.1 | -2.5889623 | 2.0767884 | |
| RF55_8207 | XP_016767693.1 | -2.6776356 | 1.9612772 | |
| RF55_3129 | GB53974 | probable RNA helicase armi | -2.4754839 | 2.1042263 |
| RF55_8822 | GB41970 | ras-like protein 2-like | -2.5012521 | 2.0767873 |
| RF55_9449 | GB55840 | protein sidekick-1-like | -1.9739288 | 2.4747175 |
| RF55_583 | GB41366 | protein MLP1-like | -2.5149738 | 1.9248983 |
| RF55_5597 | GB46705 | muscle M-line assembly protein unc-89 | -4.2010432 | 0.1942554 |
| RF55_7779 | GB40928 | tripartite motif-containing protein 2-like | -1.9058556 | 2.4627030 |
| RF55_1209 | XP_016766933.1 | -1.8405716 | 2.4685968 | |
| RF55_2958 | XP_016771468.1 | -2.2465795 | 1.9449655 | |
| RF55_7227 | GB45211 | troponin C type I | -3.4951202 | 0.6960651 |
| RF55_15397 | XP_016766611.1 | -2.1887540 | 1.9674339 | |
| RF55_3792 | GB42024 | translation initiation factor 2 | -2.3474211 | 1.7699710 |
| XLOC_009000 | -1.9702840 | 2.1399072 | ||
| RF55_1494 | XP_016771667.1 | -1.8255253 | 2.2245942 | |
| RF55_2964 | GB45277 | multidrug resistance-associated protein 4-like | -1.9410464 | 2.0992859 |
| RF55_13040 | XP_016771370.1 | -1.7071168 | 2.3051344 | |
| XLOC_009717 | -1.8567534 | 2.1300834 | ||
| RF55_6105 | GB40496 | aftiphilin-like | -1.9290288 | 2.0409600 |
| RF55_2225 | XP_016769102.1 | -2.4576563 | 1.5031576 | |
| RF55_7166 | GB51290 | ultrabithorax | -1.8574204 | 2.0948477 |
| RF55_2340 | GB55485 | DNA methyltransferase 3 | -2.2221680 | 1.7086023 |
| RF55_5141 | GB51219 | eye-specific diacylglycerol kinase | -1.5771776 | 2.3341264 |
| RF55_3424 | GB42681 | mucin-5AC-like | -1.9129112 | 1.9909363 |
| RF55_335 | GB42666 | serine palmitoyltransferase 2-like | -2.1856978 | 1.6894768 |
| RF55_2921 | GB40416 | twist | -2.1419203 | 1.6755463 |
| RF55_1441 | GB47599 | cytoplasmic dynein 1 light intermediate chain 1 | -1.3649356 | 2.3461757 |
| XLOC_001241 | -1.9132811 | 1.7758352 | ||
| RF55_404 | GB53011 | polyphosphoinositide phosphatase | -2.1273247 | 1.5615310 |
| RF55_11430 | XP_016769030.1 | -2.0923206 | 1.5691347 | |
| RF55_1609 | GB40975 | gamma-aminobutyric acid receptor subunit beta | -1.9326037 | 1.6691322 |
| RF55_5498 | GB55971 | palmitoyltransferase ZDHHC9-like | -1.8138672 | 1.6963432 |
| RF55_13282 | GB54319 | synaptotagmin 20 | -1.8203872 | 1.6560353 |
| RF55_4134 | XP_016771187.1 | -1.6338628 | 1.7842745 | |
| RF55_3869 | GB51413 | C3 and PZP-like alpha-2-macroglobulin domain-containing protein 8-like | -1.8352815 | 1.5643281 |
| RF55_5507 | XP_016772718.1 | -1.5504913 | 1.7564732 | |
| RF55_3824 | XP_016768669.1 | -1.5248764 | 1.6085396 | |
| XLOC_005436 | -1.7586500 | 1.3642664 | ||
| RF55_10519 | GB54446 | arginine kinase | -2.9654802 | 0.1456319 |
| RF55_310 | 100576851 | ornithine decarboxylase antizyme 1-like | -2.1712838 | 0.4835235 |
| RF55_3137 | GB54056 | serine hydroxymethyltransferase, cytosolic | -0.5039290 | 0.3060241 |
| RF55_6300 | XP_016767697.1 | -0.1938198 | 0.3975841 | |
| RF55_4024 | XP_016767817.1 | -0.2959092 | 0.2953227 |
Overlap of specific pheromone-sensitive alternatively-spliced genes
lf.alt <- lf.alt.splice$Name[lf.alt.splice$Name != " "] # Exclude genes with no A. mellifera names
ln.alt <- ln.alt.splice$Name[ln.alt.splice$Name != " "]
venn(list(Apis = am.alt.splice$Name,`L. flavus` = lf.alt,`L. niger` = ln.alt))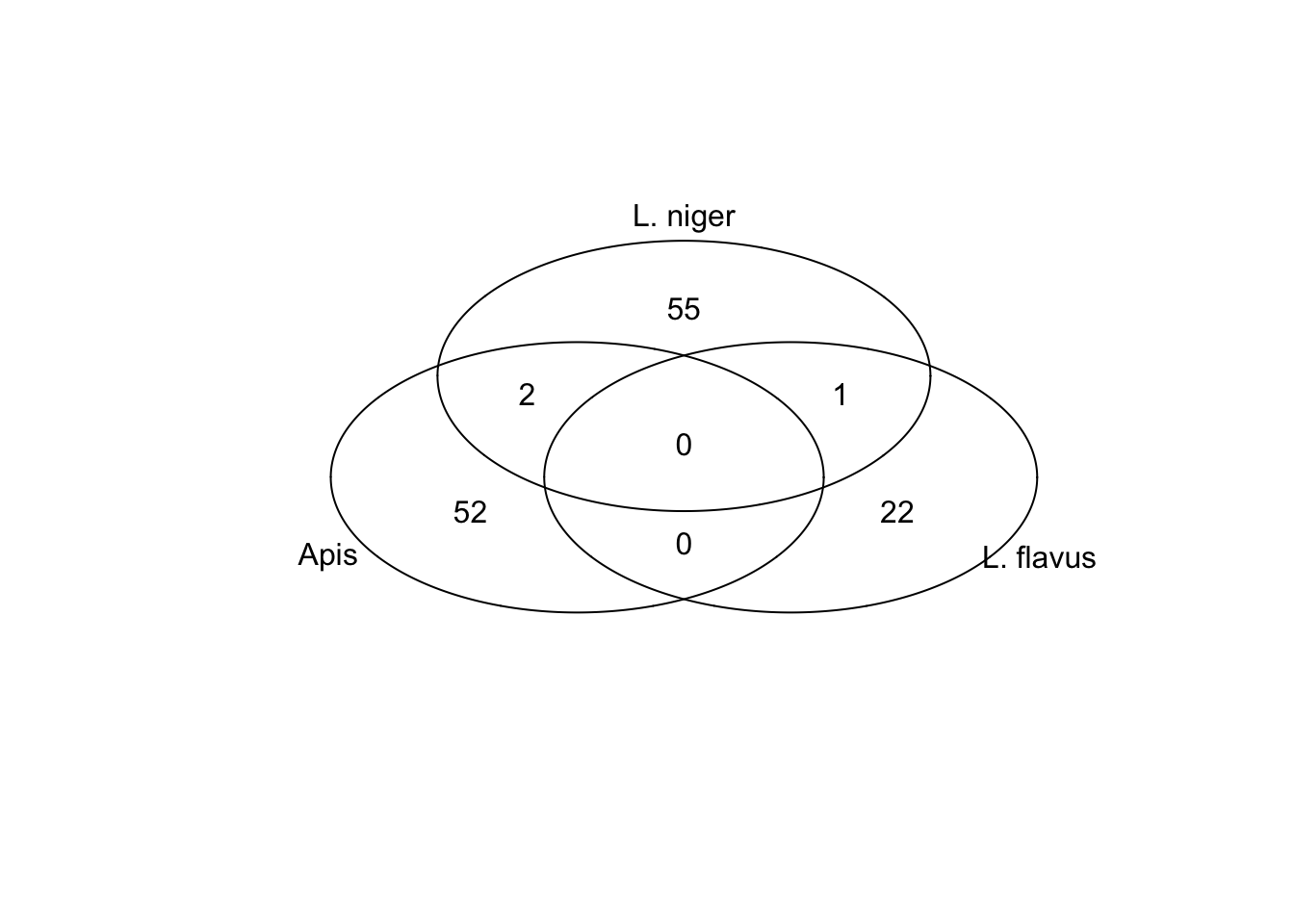
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Gene co-expression network analysis
First, we define a series of functions for gene co-expression network analysis.
# This function uses the ComBat function from the package 'sva' to remove variance in gene expression
# that is due to colony and species, allowing us to detect variance due to queen pheromone treatment.
# The use of ComBat in this fashion follows recommendations from the author of the WGCNA package.
remove.effects.combat <- function(expression.data){
sampleIDs <- rownames(expression.data)
ids <- with(treatments[match(sampleIDs, treatments$id), ],
data.frame(
id = sampleIDs,
species = species,
treatment = treatment,
colony = paste(species, colony, sep = "")))
modcombat <- model.matrix(~as.factor(treatment), data=ids)
shh <- capture.output(expression.data <- ComBat(dat = t(expression.data),
batch = ids$species,
mod = modcombat,
par.prior = TRUE))
shh <- capture.output(expression.data <- t(ComBat(dat = expression.data,
batch = ids$colony,
mod = modcombat,
par.prior = TRUE)))
list(expression.data, ids)
}
# Build a gene coexpresison network using WGCNA package
build.network <- function(expression.data.list){
# Pick the soft thresholding power that gives a model fit of R^2 > 0.8 for the scale-free topology model
soft.power <- pickSoftThreshold(expression.data.list[[1]],
RsquaredCut = 0.8, verbose = 0, powerVector = 1:30)
# Use this power to generate a gene co-expression network, using the default settings
network <- blockwiseModules(expression.data.list[[1]],
power = soft.power$powerEstimate,
networkType = "signed",
minModuleSize = 30,
verbose = 0,
saveTOMs = T)
list(network, expression.data.list[[2]])
}
# By default, WGCNA gives the transcriptional modules random names like 'turqoise' or 'darkred'.
# I think it's more helpful to define the biggest module as 'Module 1', the second biggest as 'Module 2', etc
# I use the label 'Module 0' for genes that were not assigned to a module
convert.module.colors.to.names <- function(network){
module.sizes <- table(network[[1]]$colors) %>% sort %>% rev
module.sizes <- c(module.sizes[names(module.sizes) == "grey"],
module.sizes[names(module.sizes) != "grey"])
module.mappings <- data.frame(color = names(module.sizes),
new.name = paste("Module",
0:(length(module.sizes)-1)), stringsAsFactors = F)
network[[1]]$colors <- module.mappings$new.name[match(network[[1]]$colors, module.mappings$color)]
names(network[[1]]$MEs) <- gsub("ME", "", names(network[[1]]$MEs))
names(network[[1]]$MEs) <- module.mappings$new.name[match(names(network[[1]]$MEs),
module.mappings$color)]
network
}
# Rearrange the data in a handy format for stats and plotting, and remove the 'Module 0', the un-assigned genes
rearrange.eigengene.data <- function(network.list){
cbind(network.list[[2]], network.list[[1]]$MEs) %>%
gather(Module, Eigengene, starts_with("Module")) %>%
filter(Module != "Module 0") %>%
rename(Species = species, Treatment = treatment) %>%
arrange(Species, Treatment, colony, Module)
} Make the gene co-expression network, using the set of orthologous genes for all 4 species
# The 4 bad samples get removed, then we find the orthologous genes, the data are scaled with ComBat, and then we build the network using the lowest soft-thresholding power that gives at least R^2 > 0.8 model fit
OGGs <- make.OGGs(c("am", "bt", "ln", "lf"), bad.samples = bad.samples)
network <- OGGs[[1]] %>%
remove.effects.combat() %>%
build.network() %>%
convert.module.colors.to.names()
eigen.data <- network %>% rearrange.eigengene.dataSimple statistics about the network
Here is the number of orthologous genes that form the network:
length(network[[1]]$colors)[1] 3465Here is the number and size of modules in the network - module 0 refers to the unassigned genes.
table(network[[1]]$colors) %>% pander| Module 0 | Module 1 | Module 2 | Module 3 | Module 4 | Module 5 | Module 6 |
|---|---|---|---|---|---|---|
| 107 | 1639 | 543 | 346 | 288 | 160 | 154 |
| Module 7 | Module 8 | Module 9 |
|---|---|---|
| 150 | 40 | 38 |
Make a plot of the module eigengenes
# Make a plot of the module eigengenes, split by species, module and treatment
treatments.network.plot <- function(dat){
dat %>% mutate(Treatment = replace(as.character(Treatment), Treatment == "QP", "Queen\npheromone")) %>%
ggplot(aes(Species, Eigengene, fill = Treatment)) +
geom_hline(yintercept = 0, linetype=2) +
geom_boxplot() +
facet_wrap(~Module) +
xlab(NULL) +
scale_x_discrete(labels = c("Apis\nmellifera", "Bombus\nterrestris", "Lasius\nflavus", "Lasius\nniger")) +
scale_fill_brewer(name = " ", palette = "Set3", direction = -1) +
theme_bw() +
theme(strip.background = element_blank(),
axis.text.x = element_text(face = "italic"),
panel.border = element_rect(size=0.7),
legend.position = "top")
}
eigen.data %>% treatments.network.plot()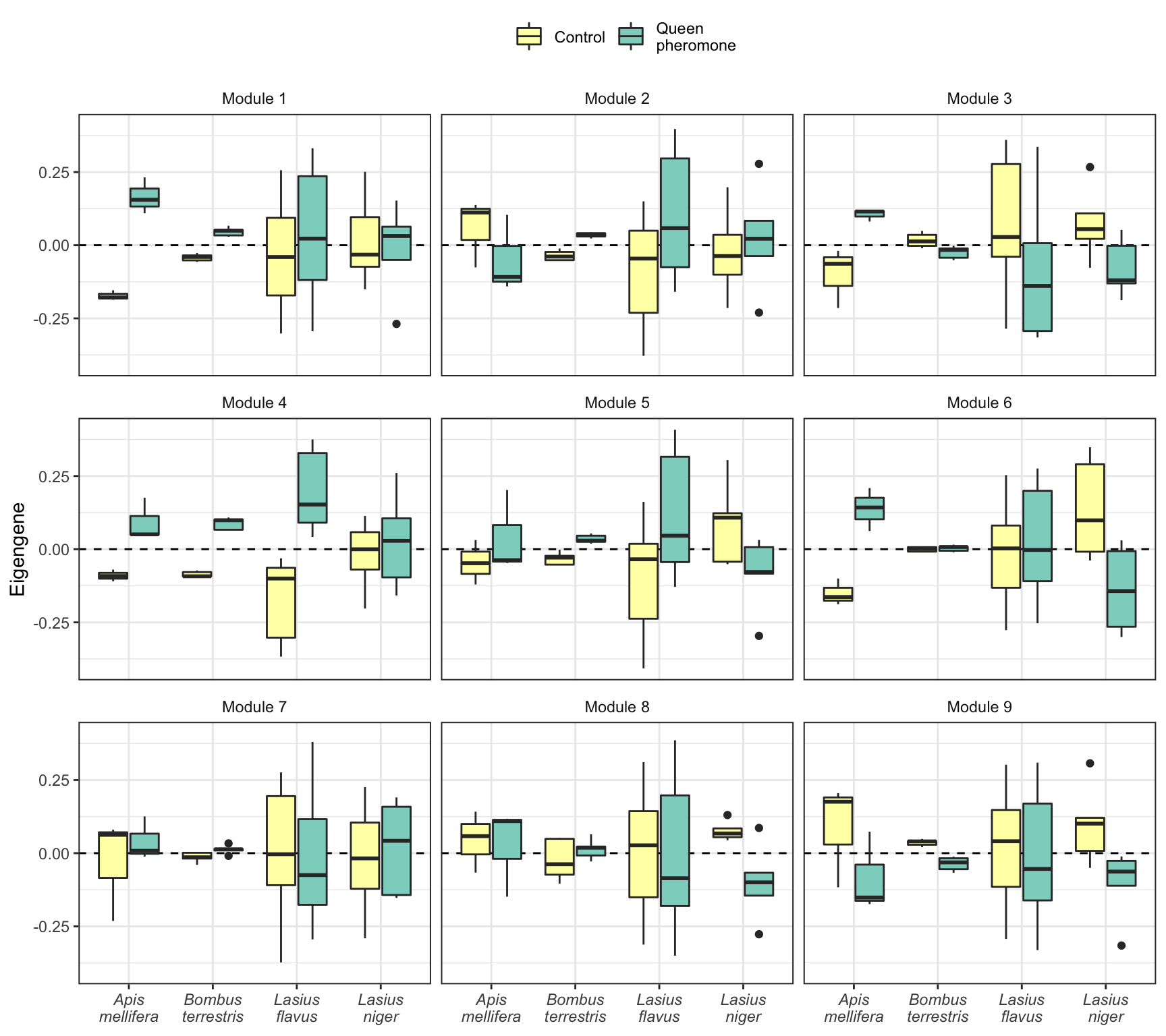
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
ggsave(eigen.data %>% treatments.network.plot(),
file = "figures/Figure 3 - Module eigengenes.pdf", height = 6.6, width = 8)
Figure 3: The figure shows the distribution of module eigengenes for each combination of module, species, and queen pheromone (QP) treatment. Positive values mean that the focal group has higher eigengenes, which derived from the relative expression levels of a module of genes, than the average.
Run multivariate Bayesian model testing for treatment and species effects on eigengenes
Since the computation time is long, the model was defined and run in a separate R script, Script to run multivariate brms model.R. It is a multivariate model with 9 response variables: the eigengenes of the nine modules. We fit colony origin as a random effect in all models, accounting for similarity in gene expression between workers from the same colony. As shown in Table S14, we ran five models of this form, with different fixed effects (i.e. models with and without the predictors Species, Treatment, and their interaction). We then calculated the posterior model probabilities for these five models using bridge sampling (which is conceptually similar to pickoing the best-fitting model by AIC). These probabilities give the chance that each model is the best-fitting one in the set being considered, given the data and the priors. The table shows that a model containing the treatment effect, but not the species effect, is the best fitting (posterior probability > 0.99), and so we conclude that pheromone treatment affects gene expression, but we have no evidence for inter-species differences, or a treatment-by-species interaction.
Table S14: The posterior model probabilities of five competing multivariate Bayesian models of the module eigengene dataset. The best-fitting model (with posterior probability of almost 1) contains the treatment effect only (not the species effect, or the treatment-by-species interaction).
readRDS("data/brms_model_comparisons.rds") %>% pander()| Posterior model probability | |
|---|---|
| Treatment x Species | 0 |
| Treatment + Species | 0 |
| Treatment | 1 |
| Species | 0 |
| Intercept only | 0 |
Table S15: Full summary of the best-fitting multivariate Bayesian model of the eigengene data for all nine modules, implemented in the programming language Stan via the R package brms. The most salient part of the output is the population-level effects (often called fixed effects), which give the coefficients for the intercept and the effect of queen pheromone treatment on the eigengenes for each module. The 9 response variables were all scaled to have mean 0 and variance 1 before running the model, meaning that the estimates can be interpreted as Cohen’s \(d\) effect size. The remaining sections describe the (co)variance associated with colony (which appears to be low), and the covariance in the residuals (which illustrates how eigengenes are correlated across modules).
cat(paste(readLines("data/brms_model.txt"), "\n", sep = "")) Family: MV(gaussian, gaussian, gaussian, gaussian, gaussian, gaussian, gaussian, gaussian, gaussian)
Links: mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
Formula: m1 ~ Treatment + (1 | p | colony)
m2 ~ Treatment + (1 | p | colony)
m3 ~ Treatment + (1 | p | colony)
m4 ~ Treatment + (1 | p | colony)
m5 ~ Treatment + (1 | p | colony)
m6 ~ Treatment + (1 | p | colony)
m7 ~ Treatment + (1 | p | colony)
m8 ~ Treatment + (1 | p | colony)
m9 ~ Treatment + (1 | p | colony)
Data: eigen.data %>% mutate(Module = gsub("Module ", "m" (Number of observations: 39)
Samples: 4 chains, each with iter = 5000; warmup = 2500; thin = 1;
total post-warmup samples = 10000
Group-Level Effects:
~colony (Number of levels: 27)
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
sd(m1_Intercept) 0.08 0.06 0.00 0.21 4356 1.00
sd(m2_Intercept) 0.04 0.03 0.00 0.12 7092 1.00
sd(m3_Intercept) 0.04 0.03 0.00 0.13 10000 1.00
sd(m4_Intercept) 0.04 0.03 0.00 0.13 8519 1.00
sd(m5_Intercept) 0.05 0.03 0.00 0.12 4317 1.00
sd(m6_Intercept) 0.05 0.04 0.00 0.14 4690 1.00
sd(m7_Intercept) 0.06 0.04 0.00 0.17 10000 1.00
sd(m8_Intercept) 0.13 0.09 0.01 0.34 6666 1.00
sd(m9_Intercept) 0.04 0.03 0.00 0.12 5685 1.00
cor(m1_Intercept,m2_Intercept) 0.02 0.32 -0.58 0.60 10000 1.00
cor(m1_Intercept,m3_Intercept) 0.01 0.32 -0.60 0.61 10000 1.00
cor(m2_Intercept,m3_Intercept) -0.01 0.32 -0.62 0.60 10000 1.00
cor(m1_Intercept,m4_Intercept) -0.00 0.32 -0.62 0.61 10000 1.00
cor(m2_Intercept,m4_Intercept) 0.05 0.32 -0.57 0.64 10000 1.00
cor(m3_Intercept,m4_Intercept) -0.02 0.32 -0.62 0.60 10000 1.00
cor(m1_Intercept,m5_Intercept) 0.02 0.32 -0.58 0.61 10000 1.00
cor(m2_Intercept,m5_Intercept) 0.00 0.32 -0.60 0.59 10000 1.00
cor(m3_Intercept,m5_Intercept) -0.02 0.32 -0.63 0.59 10000 1.00
cor(m4_Intercept,m5_Intercept) 0.02 0.32 -0.58 0.62 10000 1.00
cor(m1_Intercept,m6_Intercept) 0.00 0.31 -0.59 0.60 10000 1.00
cor(m2_Intercept,m6_Intercept) -0.02 0.32 -0.61 0.59 10000 1.00
cor(m3_Intercept,m6_Intercept) 0.05 0.32 -0.58 0.64 10000 1.00
cor(m4_Intercept,m6_Intercept) 0.03 0.31 -0.57 0.63 10000 1.00
cor(m5_Intercept,m6_Intercept) 0.03 0.32 -0.58 0.63 10000 1.00
cor(m1_Intercept,m7_Intercept) -0.01 0.32 -0.61 0.59 10000 1.00
cor(m2_Intercept,m7_Intercept) 0.02 0.32 -0.59 0.62 10000 1.00
cor(m3_Intercept,m7_Intercept) 0.06 0.32 -0.57 0.65 10000 1.00
cor(m4_Intercept,m7_Intercept) 0.02 0.32 -0.58 0.62 10000 1.00
cor(m5_Intercept,m7_Intercept) -0.01 0.32 -0.61 0.59 10000 1.00
cor(m6_Intercept,m7_Intercept) -0.02 0.32 -0.63 0.59 10000 1.00
cor(m1_Intercept,m8_Intercept) 0.01 0.31 -0.59 0.61 10000 1.00
cor(m2_Intercept,m8_Intercept) -0.01 0.31 -0.60 0.58 10000 1.00
cor(m3_Intercept,m8_Intercept) 0.04 0.32 -0.57 0.64 10000 1.00
cor(m4_Intercept,m8_Intercept) 0.02 0.31 -0.58 0.61 10000 1.00
cor(m5_Intercept,m8_Intercept) -0.00 0.32 -0.60 0.60 10000 1.00
cor(m6_Intercept,m8_Intercept) -0.04 0.31 -0.62 0.57 7970 1.00
cor(m7_Intercept,m8_Intercept) -0.04 0.32 -0.62 0.58 6458 1.00
cor(m1_Intercept,m9_Intercept) -0.01 0.32 -0.62 0.60 10000 1.00
cor(m2_Intercept,m9_Intercept) 0.05 0.33 -0.59 0.65 10000 1.00
cor(m3_Intercept,m9_Intercept) -0.00 0.32 -0.61 0.60 10000 1.00
cor(m4_Intercept,m9_Intercept) -0.05 0.32 -0.64 0.57 10000 1.00
cor(m5_Intercept,m9_Intercept) 0.04 0.31 -0.57 0.62 10000 1.00
cor(m6_Intercept,m9_Intercept) 0.02 0.31 -0.59 0.62 8255 1.00
cor(m7_Intercept,m9_Intercept) 0.01 0.32 -0.60 0.61 6350 1.00
cor(m8_Intercept,m9_Intercept) 0.03 0.32 -0.58 0.63 6231 1.00
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
m1_Intercept -0.27 0.18 -0.64 0.09 4403 1.00
m2_Intercept -0.23 0.19 -0.60 0.14 5015 1.00
m3_Intercept 0.23 0.19 -0.16 0.60 5448 1.00
m4_Intercept -0.63 0.15 -0.92 -0.32 5864 1.00
m5_Intercept -0.18 0.18 -0.53 0.19 4295 1.00
m6_Intercept 0.03 0.20 -0.36 0.43 5752 1.00
m7_Intercept -0.05 0.20 -0.46 0.34 5803 1.00
m8_Intercept 0.13 0.21 -0.28 0.54 5686 1.00
m9_Intercept 0.32 0.17 -0.01 0.67 4355 1.00
m1_TreatmentQP 0.56 0.26 0.04 1.06 4299 1.00
m2_TreatmentQP 0.48 0.27 -0.06 1.00 4542 1.00
m3_TreatmentQP -0.47 0.28 -1.01 0.10 5341 1.00
m4_TreatmentQP 1.28 0.22 0.85 1.71 5443 1.00
m5_TreatmentQP 0.37 0.26 -0.15 0.86 4059 1.00
m6_TreatmentQP -0.07 0.29 -0.65 0.50 5358 1.00
m7_TreatmentQP 0.11 0.30 -0.47 0.69 5655 1.00
m8_TreatmentQP -0.27 0.29 -0.85 0.32 5407 1.00
m9_TreatmentQP -0.66 0.25 -1.16 -0.17 4120 1.00
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
sigma_m1 0.82 0.08 0.69 0.98 10000 1.00
sigma_m2 0.84 0.08 0.70 1.01 7447 1.00
sigma_m3 0.87 0.09 0.72 1.05 7227 1.00
sigma_m4 0.70 0.07 0.58 0.85 10000 1.00
sigma_m5 0.82 0.07 0.69 0.97 5832 1.00
sigma_m6 0.90 0.09 0.75 1.10 6538 1.00
sigma_m7 0.92 0.09 0.76 1.12 10000 1.00
sigma_m8 0.91 0.10 0.75 1.12 10000 1.00
sigma_m9 0.79 0.07 0.66 0.94 6247 1.00
Residual Correlations:
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
rescor(m1,m2) 0.56 0.10 0.35 0.73 6438 1.00
rescor(m1,m3) 0.69 0.08 0.51 0.82 7120 1.00
rescor(m2,m3) 0.25 0.12 -0.00 0.48 10000 1.00
rescor(m1,m4) 0.53 0.10 0.31 0.71 8099 1.00
rescor(m2,m4) 0.73 0.07 0.56 0.84 10000 1.00
rescor(m3,m4) 0.10 0.13 -0.17 0.35 10000 1.00
rescor(m1,m5) 0.59 0.10 0.38 0.76 5667 1.00
rescor(m2,m5) 0.66 0.08 0.48 0.79 5941 1.00
rescor(m3,m5) 0.42 0.11 0.19 0.61 8145 1.00
rescor(m4,m5) 0.71 0.07 0.55 0.83 7301 1.00
rescor(m1,m6) 0.65 0.09 0.45 0.79 6974 1.00
rescor(m2,m6) 0.19 0.13 -0.07 0.43 10000 1.00
rescor(m3,m6) 0.64 0.08 0.46 0.78 8022 1.00
rescor(m4,m6) 0.32 0.12 0.06 0.54 8027 1.00
rescor(m5,m6) 0.78 0.06 0.65 0.87 10000 1.00
rescor(m1,m7) 0.59 0.10 0.36 0.75 10000 1.00
rescor(m2,m7) 0.59 0.09 0.39 0.74 10000 1.00
rescor(m3,m7) 0.71 0.07 0.54 0.83 10000 1.00
rescor(m4,m7) 0.32 0.12 0.06 0.55 10000 1.00
rescor(m5,m7) 0.35 0.12 0.11 0.56 8081 1.00
rescor(m6,m7) 0.21 0.13 -0.07 0.45 8440 1.00
rescor(m1,m8) 0.53 0.11 0.29 0.71 10000 1.00
rescor(m2,m8) 0.53 0.10 0.30 0.71 10000 1.00
rescor(m3,m8) 0.55 0.10 0.34 0.72 10000 1.00
rescor(m4,m8) 0.46 0.11 0.22 0.66 10000 1.00
rescor(m5,m8) 0.49 0.11 0.26 0.68 10000 1.00
rescor(m6,m8) 0.39 0.12 0.13 0.60 10000 1.00
rescor(m7,m8) 0.44 0.12 0.18 0.65 10000 1.00
rescor(m1,m9) 0.57 0.10 0.35 0.74 5328 1.00
rescor(m2,m9) 0.77 0.06 0.64 0.87 7273 1.00
rescor(m3,m9) 0.54 0.10 0.33 0.70 10000 1.00
rescor(m4,m9) 0.45 0.11 0.21 0.65 10000 1.00
rescor(m5,m9) 0.83 0.05 0.71 0.90 10000 1.00
rescor(m6,m9) 0.60 0.09 0.39 0.75 10000 1.00
rescor(m7,m9) 0.57 0.10 0.35 0.73 10000 1.00
rescor(m8,m9) 0.52 0.11 0.28 0.70 10000 1.00
Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
is a crude measure of effective sample size, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).Plot the correlations between all modules and the pheromone treatment
meta.module.plot <- function(network){
MET <- network[[1]]$MEs
MET <- data.frame(QP = (network[[2]]$treatment %>% as.numeric())-1, MET) %>% dplyr::select(-Module.0)
names(MET) <- gsub("[.]", " ", names(MET))
cluster <- (1 - cor(MET)) %>% as.dist() %>% hclust()
ordering <- cluster$labels[cluster$order]
heat.map.data <- cor(MET) %>% melt %>%
mutate(Var1 = factor(Var1, levels = ordering),
Var2 = factor(Var2, levels = ordering)) %>%
rename(Corr = value)
heat.map <- heat.map.data %>% ggplot(aes(Var1, Var2, fill = Corr)) + geom_tile() +
scale_fill_gradient2(low = brewer.pal(9, "RdBu")[8],
mid = "white",
high = brewer.pal(9, "RdBu")[2]) +
xlab(NULL) + ylab(NULL) +
theme_bw() +
theme(panel.border = element_blank(),
panel.grid = element_blank()) +
scale_x_discrete(expand = c(0,0)) +
scale_y_discrete(expand = c(0,0))
dendrogram <- ggdendrogram(cluster) + theme(axis.text.x = element_blank(), axis.text.y = element_blank())
p1 <- grid.arrange(dendrogram, heat.map)
invisible(p1)
}
meta.module.plot(network)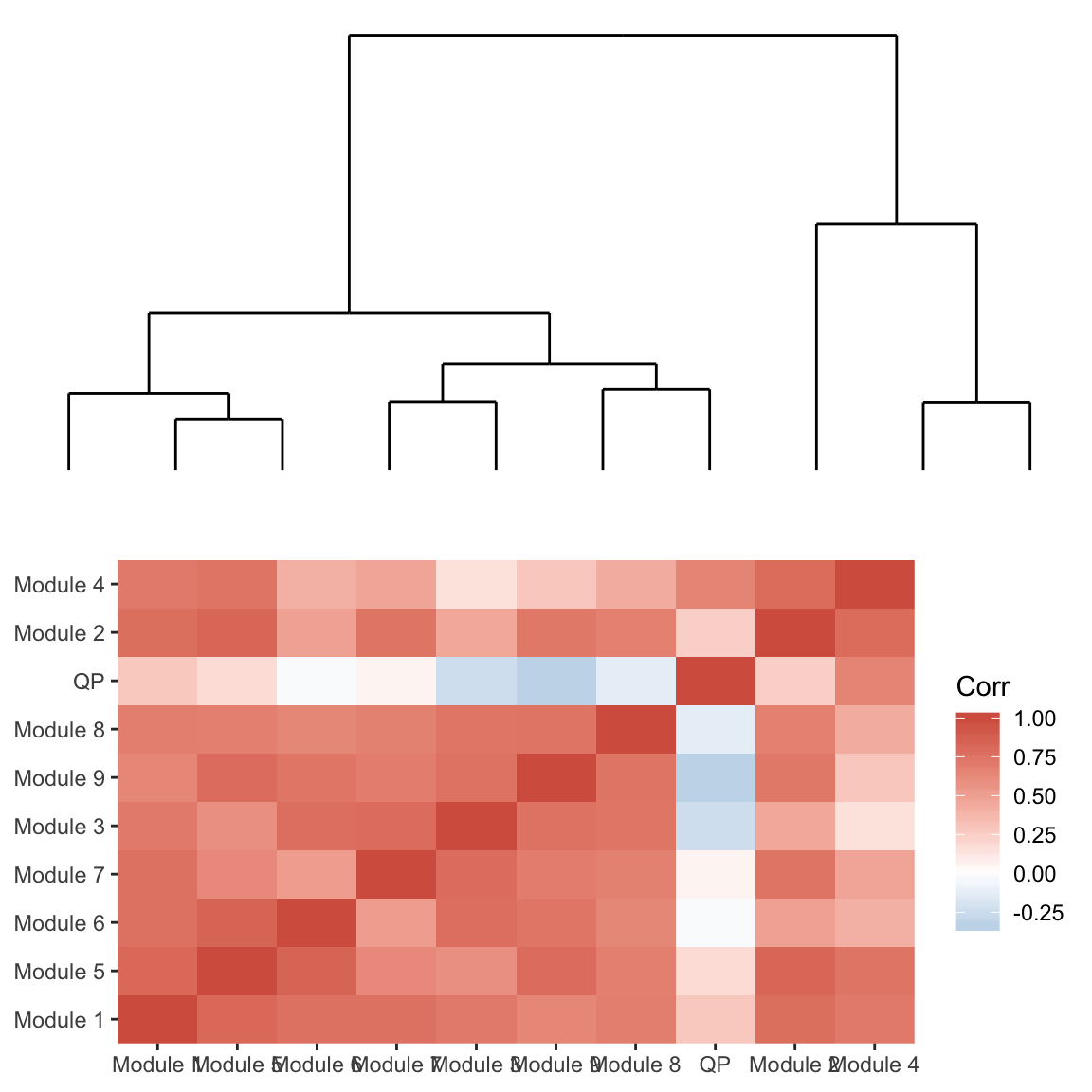
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Dendrogram and heat map showing the correlations among module eigengene values and the queen pheromone treatment (QP; coded as zero and 1 for the control and treatment respectively). Modules with red colour, or which are close on the dendrogram, show more correlated expression. The queen pheromone treatment was correlated with Module 5, but was relatively uncorrelated with the other modules.
Table S16: Results of Spearman’s rank correlations testing for a relationship between the effect of queen pheromone on gene expression, and the connectedness of the gene. Negative values of Spearman’s Rho mean that highly pheromone-sensitive genes tend to have lower connectedness.
load("blockwiseTOM-block.1.RData") # Load the TOM from the network analysis - can be used to find connectedness for each ortholog
# Get the phermonone sensitivity value for each of the OGGs, and line it up with the connectedness data
suppressMessages(dd <- OGGs[[2]] %>% left_join(tbl(my_db, "ebseq_gene_am") %>% select(gene, PostFC) %>% rename(am=gene), copy=T) %>% rename(am_FC = PostFC) %>%
left_join(tbl(my_db, "ebseq_gene_bt") %>% select(gene, PostFC) %>%
rename(bt=gene), copy=T) %>% rename(bt_FC = PostFC) %>%
left_join(tbl(my_db, "ebseq_gene_lf") %>% select(gene, PostFC) %>%
rename(lf=gene), copy=T) %>% rename(lf_FC = PostFC) %>%
left_join(tbl(my_db, "ebseq_gene_ln") %>% select(gene, PostFC) %>%
rename(ln=gene), copy=T) %>% rename(ln_FC = PostFC) %>%
mutate(k = colSums(as.matrix(TOM))))
do.spearman <- function(dd, species, name){ # run spearman on each species
test <- cor.test(dd$k, dd[, names(dd) == species] %>% log2 %>% abs, method = "spearman") # Note that we convert the fold-change in expression with abs(log2(x)), to get pheromone sensitivity
with(test, data.frame(Species = name, rho = estimate, p = p.value, row.names = NULL))
}
suppressWarnings(results <- rbind(do.spearman(dd, "am_FC", "Apis mellifera"),
do.spearman(dd, "bt_FC", "Bombus terrestris"),
do.spearman(dd, "lf_FC", "Lasius flavus"),
do.spearman(dd, "ln_FC", "Lasius niger")))
rownames(results) <- NULL
saveRDS(results, file = "supplement/tab_S16.rds")
pander(results)| Species | rho | p |
|---|---|---|
| Apis mellifera | -0.2463 | 4.834e-49 |
| Bombus terrestris | -0.2988 | 2.076e-72 |
| Lasius flavus | -0.4151 | 1.854e-144 |
| Lasius niger | -0.3159 | 4.677e-81 |
Characteristics of pheromone-sensitive genes in Apis
In this section, we search for correlates of the absolute Log_2 fold change in response to pheromone (where positive values denote genes whose expression differs strongly between the control and pheromone treatment). This section makes use of data kindly passed to us by Soojin Yi and Brendan Hunt.
The data on queen and worker-specific gene expression come from Grozinger et al. 2007. We found that pheromone-sensitive genes tend to be over-expressed by queens relative to sterile workers. However, genes that are over-expressed by fertile workers relative to sterile workers did not tend to be more (or less) pheromone-sensitive.
The methylation level (i.e. % methylated cytosines) data come from Galbraith et al. 2016 (provided by Soojin Yi). We found a negative correlation between methylation and pheromone-sensitivity, suggesting that pheromone-sensitive genes are hypomethylated.
The CpG O/E values were calculated by Yi and colleagues for the latest A. mellifera genome annotation (as of late 2017). We found a positive correlation between CpG O/E and pheromone-sensitivity. High CpG is associated with lower rate of DNA methylation, again suggesting that pheromone-sensitive genes are hypomethylated.
The columns on the H3K4me3, H3K27ac, and H3K36me3 histon modifications were calculated from the raw data from Wojciechowski et al. 2018, using the code in the separate R script
wojciechowski_histone_analysis.R.The estimates of \(\gamma\), a measure of positive and negative selection similar to dN/dS, come from Harpur et al. 2014 PNAS. There was a significant positive correlation between \(\gamma\) and sensitivity to queen pheromone, suggesting that highly pheromone-sensitive genes tend to be positively selected.
The codon adaptation index was provided by Brendan Hunt. A high codon adaptation index denotes high codon usage bias, meaning that certain synonymous codons are more common than others. Pheromone-sensitive genes showed low codon usage bias.
Lastly, ‘expression level’ refers to the average expression of each gene, expressed as TPM (transcripts per million) as measured by the software RSEM in the present study. Highly expressed genes tended to be less pheromone-sensitive.
# import and clean up the data provided by Brendan Hunt
hunt.data <- read.delim("data/apis_gene_comparisons/Amel_AllData_012709.txt",
header=T, stringsAsFactors = FALSE) %>%
mutate(log2RW.SW = log2(RW_bagel / SW_bagel)) # calculate log fold difference in gene expression between fertile and sterile workers
entrez.tbl <- read.delim("data/apis_gene_comparisons/am.gene_info.txt", stringsAsFactors = FALSE)[,c(2,5,6)] # import table of gene names (Entrez, old Beebase, and new Beebase)
names(entrez.tbl) <- c("entrez.id", "beebase1", "beebase2")
entrez.tbl <- entrez.tbl %>% mutate(beebase2 = gsub("BEEBASE:", "", beebase2))
entrez.tbl$beebase2[entrez.tbl$beebase2 == "-"] <- entrez.tbl$beebase1[entrez.tbl$beebase2 == "-"]
entrez.tbl$beebase2[grep("\\|", entrez.tbl$beebase2)] <- unname(unlist(sapply(entrez.tbl$beebase2[grep("\\|", entrez.tbl$beebase2)], function(x){
namess <- strsplit(x, split = "\\|")[[1]]
hits <-str_detect(namess, "GB")
if(sum(hits) == 0) return(NA)
return(namess[hits])
})))
hunt.data <- hunt.data %>%
dplyr::select(ID, log2Q.SW, log2RW.SW, CAI, cpgOE) %>% # get gene ID and the relevant data
rename(beebase1 = ID) %>% # merge based on beebase IDs
left_join(entrez.tbl, by = "beebase1") %>%
filter(!is.na(beebase2) & beebase2 != "-") %>%
rename(gene = beebase2) %>%
dplyr::select(gene, log2Q.SW, log2RW.SW, CAI, cpgOE)
hunt.data <- left_join(tbl(my_db, "ebseq_gene_am") %>% # merge Hunt's data with our phermone sensitivity data
dplyr::select(gene, PostFC) %>% collect(), hunt.data, by = "gene") %>%
left_join(tbl(my_db, "bee_names") %>% collect, by = "gene") # also add the gene names
# Import methylation data provided by Soojin Yi and Xin Wu (teh data are from Galbraith et al PNAS)
methylation <- read.csv("data/apis_gene_comparisons/apis_gene_methyl_CG_OE.csv", stringsAsFactors = FALSE)
methylation <- tbl(my_db, "ebseq_gene_am") %>% collect %>% left_join(methylation, by = "gene") %>%
filter(!is.na(Gene_body_methylation)) %>%
left_join(tbl(my_db, "bee_names") %>% collect(), by = "gene") %>% distinct(gene, .keep_all = T)
# Import gamma data from Harpur et al 2014 PNAS
gamma_am <- tbl(my_db, "ebseq_gene_am") %>% collect() %>%
left_join(read.table("data/apis_gene_comparisons/harpur_etal_gamma.txt", header=TRUE, stringsAsFactors=FALSE) %>% rename(gene = Gene), by = "gene") %>%
filter(!is.na(gamma)) %>%
dplyr::select(gene, PostFC, gamma) %>%
mutate(PostFC = log2(PostFC)) %>% left_join(tbl(my_db, "bee_names"), copy=TRUE, by = "gene") %>%
arrange(-abs(PostFC))
merged <- hunt.data %>% dplyr::select(gene, PostFC, log2Q.SW, log2RW.SW, CAI) %>%
left_join(methylation %>% dplyr::select(gene, CG_OE, Gene_body_methylation), by = "gene") %>% # merge in methylation data provided by Soojin Yi
left_join(gamma_am %>% dplyr::select(gene, gamma), by = "gene") %>% # merge in gamma data from Harpur et al
left_join(data.frame(gene = tbl(my_db, "rsem_am") %>%
dplyr::select(gene) %>% collect(), # calculate overall expression level from our own data
expression.level = (tbl(my_db, "rsem_am") %>%
dplyr::select(-gene) %>%
collect %>% rowSums())), by = "gene") %>%
left_join(dd %>% dplyr::select(am, k) %>% rename(gene=am), by = "gene") %>%
mutate(CG_OE = -log2(CG_OE), # log2 CpG O/E ratio - change the sign, so that high values mean high methylation
expression.level = log10(expression.level), # log10 the expression level
PostFC = abs(log2(PostFC))) %>% # absolute log2 fold-change in response to pheromones (i.e. 'pheromone sensitivity' score)
as.data.frame() #%>%
# dplyr::select(-gene)
# Add the data from Wojciechowski et al. 2018 Genome Biology
# There is gene-level data on the caste difference in 3 different histone modifications (from ChIP-seq),
# These data were created from Wojciechowski et al.'s raw data, as described in the separate R script wojciechowski_histone_analysis.R
merged <- merged %>%
left_join(tbl(my_db, "H3K4me3") %>% collect() %>% mutate(H3K4me3_caste = caste_difference) %>% select(gene, H3K4me3_caste), by = "gene") %>%
left_join(tbl(my_db, "H3K27ac") %>% collect() %>% mutate(H3K27ac_caste = caste_difference) %>% select(gene, H3K27ac_caste), by = "gene") %>%
left_join(tbl(my_db, "H3K36me3") %>% collect() %>% mutate(H3K36me3_caste = caste_difference) %>% select(gene, H3K36me3_caste), by = "gene") %>%
dplyr::select(-gene)
m.rename <- function(merged, col, new) {names(merged)[names(merged) == col] <- new; merged}
merged <- merged %>% m.rename("PostFC", "Pheromone sensitivity\n(absolute log fold)") # re-label all the variables nicely
merged <- merged %>% m.rename("log2Q.SW", "Upregulation in adult queens\nover sterile workers (log fold)")
merged <- merged %>% m.rename("log2RW.SW", "Upregulation in fertile workers\nover sterile workers (log fold)")
merged <- merged %>% m.rename("CAI", "Codon usage bias\n(CAI)")
merged <- merged %>% m.rename("CG_OE", "DNA methylation frequency\n(CpG depletion)")
merged <- merged %>% m.rename("Gene_body_methylation", "DNA methylation frequency\n(BiS-seq)")
merged <- merged %>% m.rename("gamma", "Positive selection\n(Gamma)")
merged <- merged %>% m.rename("expression.level", "Log Expression level")
merged <- merged %>% m.rename("k", "Connectivity in the\ntranscriptome")
merged <- merged %>% m.rename("H3K4me3_caste", "Caste difference in\nH3K4me3 modification")
merged <- merged %>% m.rename("H3K27ac_caste", "Caste difference in\nH3K27ac modification")
merged <- merged %>% m.rename("H3K36me3_caste", "Caste difference in\nH3K36me3 modification")
entrez.tbl <- entrez.tbl %>% rename(gene = beebase2) %>% select(gene, entrez.id)
# function to reorder a correlation matrix using hierarchical clustering
reorder_cormat <- function(cormat){
dd <- as.dist((1-cormat)/2)
hc <- hclust(dd)
cormat[hc$order, hc$order]}
cormat <- reorder_cormat(cor(merged, use = 'pairwise.complete.obs', method = "spearman"))
cormat[upper.tri(cormat)] <- NA
diag(cormat) <- NA
cor.matrix <- (melt(cormat) %>% filter(!is.na(value)))[,1:2] %>% mutate(cor=0,p=0)
for(i in 1:nrow(cor.matrix)) cor.matrix[i, 3:4] <- cor.test(merged[, names(merged) == cor.matrix$Var1[i]], merged[, names(merged) == cor.matrix$Var2[i]])[c(4,3)] %>% unlist
cor.matrix$p <- p.adjust(cor.matrix$p, method = "BH") # apply B-H p value correction
cor.matrix$sig <- ""
cor.matrix$sig[cor.matrix$p < 0.05] <- "*"
cor.matrix$sig[cor.matrix$p < 0.001] <- "**"
cor.matrix$sig[cor.matrix$p < 0.0001] <- "***"
cor.matrix$label <- paste(format(round(cor.matrix$cor,2), nSmall =2), cor.matrix$sig, sep = "")
correlation.plot <- cor.matrix %>%
ggplot(aes(Var1, Var2, fill=cor)) +
geom_tile(colour = "grey10", size = 0.4, linetype = 3) +
scale_fill_distiller(palette = "RdYlBu") +
geom_text(aes(label = label), colour = "grey25", size = 3.5) +
xlab(NULL) + ylab(NULL) +
scale_x_discrete(expand=c(0.01,0.01)) + scale_y_discrete(expand = c(0.01, 0.01)) +
theme_minimal() + theme(axis.text.x = element_text(angle = 45, hjust = 1,
face = c("plain", "bold", rep("plain", 5), "bold", rep("plain", 4))),
axis.text.y = element_text(face = c("bold", "plain", "bold", rep("plain", 5), "bold", "plain", "plain")),
panel.grid = element_blank(),
legend.position = "none")
ggsave(correlation.plot, file = "figures/Figure 5 - correlations.pdf", width = 8, height = 8)
correlation.plot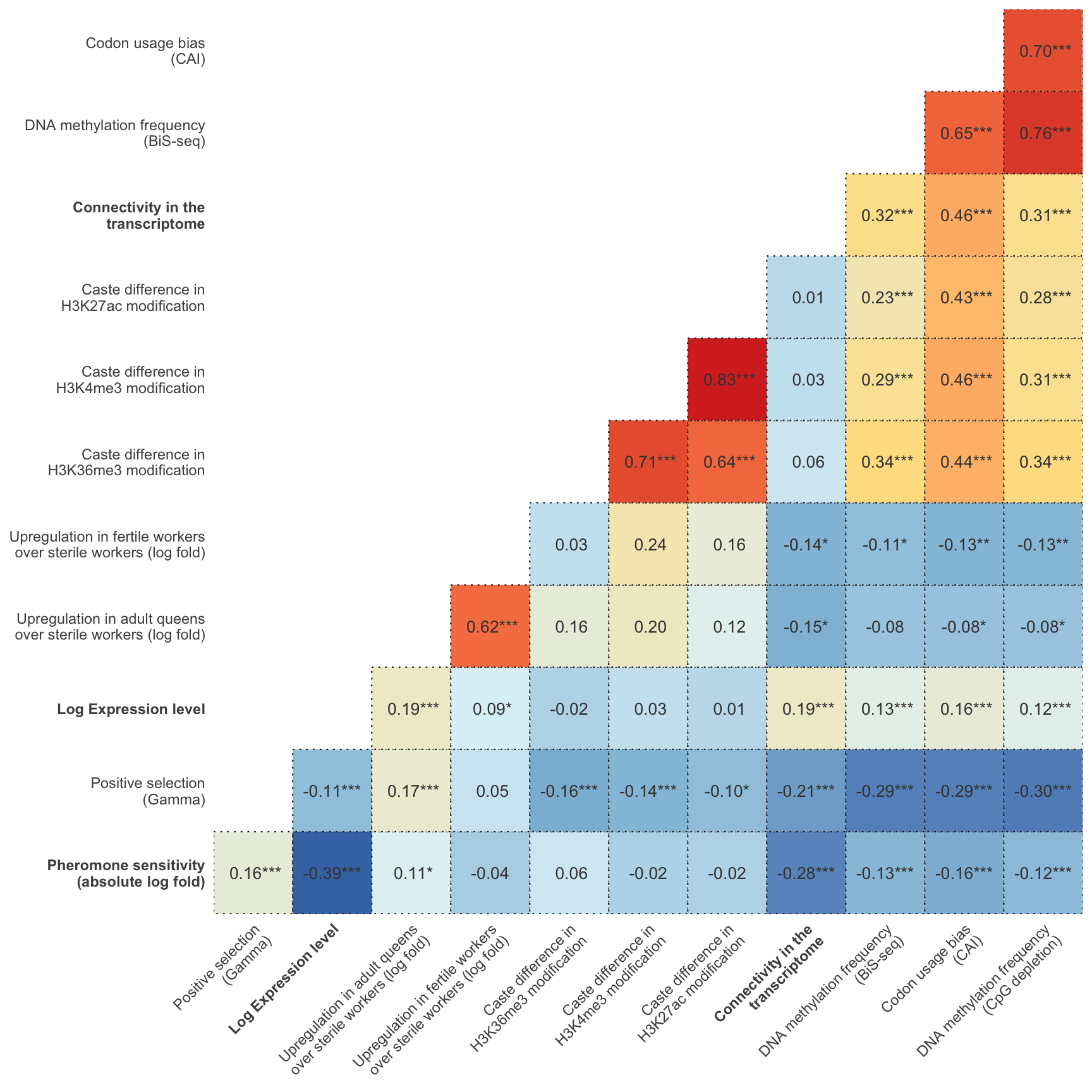
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Figure 5: Spearman correlations for various gene-level measurements from the present study and earlier research, for Apis mellifera (measurements from the present study are shown in bold). ‘Pheromone sensitivity’ was calculated as the absolute value of the Log\(_2\) fold difference in expression between pheromone treatment and the control. Expression level shows the logarithm of the average across our 6 Apis libraries. For the ‘Upregulation in queens/fertile workers’ data (Grozinger et al. 2007), positive values denote genes that have higher expression in queens or fertile workers, relative to sterile workers. For the three histone modification variables (Wojciechowski et al. 2018), high values indicate that the modification is more abundant in queen-destined larvae, and low values indicate it is more abundant in worker-destined larvae. The two DNA methylation variables give two different measures of the amount of gene body DNA methylation, namely an indirect measure (-log CpG O/E ratio) and a direct measure (BiS-seq, Galbraith et al. 2016). Codon usage bias was estimated using the codon adaptation index: high values indicate bias for particular synonymous codons. Lastly, the parameter gamma (\(\gamma\)) describes the form of selection, where positive values denote positive selection, and negative values purifying selection (data from Hunt et al. 2014).
Contrasting our Lasius data with results from Morandin et al. 2016
Morandin et al. 2016 (Genome Biology 17:43) studied whole transcriptomes from queens and workers in 16 diverse ant species, including two other species from the genus Lasius. Using BLAST, they grouped genes into OGGs (orthologous gene groups), and built a co-expression network using all the OGGs that were common to all 16 species via the software WGCNA (much like the present study). Their analysis yielded 36 modules, of which many showed significant queen-worker differences in their module eigengenes. Here, we want to test whether these queen- and worker-like modules significantly overlap with the pheromone-sensitive modules in the present study.
To do this, we used BLAST to identify orthologous gens in L. niger and L. flavus that belong to one of Morandin et al.’s OGGs. We then tested for significantly-greater-than-random overlap between Morandin et al’s modules, and our own study’s modules, using hypergeometric tests.
Of all the possible module pairs, we found 6 pairs that overlapped significantly more than expected (FDR-corrected). One of these pairs included our highly pheromone-sensitive module, Module 4, which overlapped with the caste-biased Module 13 from Morandin et al. The intersecting genes include protein take-out like, a NAD kinase 2, a serine protease, and histone H2A-like.
morandin.orthology <- read.csv("data/morandin_comparison_data/Morandin to Holman orthology.csv", stringsAsFactors = FALSE)
morandin.module.membership <- read.csv("data/morandin_comparison_data/Morandin module membership.csv", stringsAsFactors = FALSE)
morandin.module.caste.bias <- read.csv("data/morandin_comparison_data/Morandin module caste bias.csv", stringsAsFactors = FALSE)
# Create list of all the L. niger & L. flavus genes are are part of
# the Orthologous Gene Groups (OGGs) from Morandin et al.
# Here are the 3634 genes for which we have 1-to-1 orthologs in all 18 species:
morandin.oggs <- make.OGGs(c("ln", "lf"))[[2]] %>%
left_join(morandin.orthology, by = "ln") %>%
filter(!is.na(morandin.ogg)) %>%
left_join(morandin.module.membership, by = "morandin.ogg") %>%
left_join(tbl(my_db, "ln2am") %>%
left_join(tbl(my_db, "bee_names"), by = c("am" = "gene")) %>%
filter(!is.na(name)) %>% select(ln, name, am) %>%
rename(apis.name = name) %>% collect(n=Inf), by = "ln") %>%
left_join(morandin.module.caste.bias, by = "module") %>%
rename(morandin.module = module) %>%
left_join(data.frame(ln = OGGs[[2]]$ln,
holman.module = network[[1]]$colors,
stringsAsFactors = F), by = "ln") %>%
left_join(tbl(my_db, "ebseq_gene_ln") %>%
select(gene, PostFC) %>% collect(n=Inf) %>% rename(ln = gene), by = "ln") %>%
rename(FC.pheromone = PostFC)
overlaps <- table(morandin.oggs$morandin.module,
morandin.oggs$holman.module) %>%
melt() %>% rename(morandin.module = Var1,
holman.module = Var2,
overlaps = value) %>%
filter(holman.module != "Module 0") %>%
left_join(table(morandin.oggs$morandin.module) %>%
melt() %>% rename(mor.mod.size = value),
by = c("morandin.module" = "Var1")) %>%
left_join(table(morandin.oggs$holman.module) %>% melt() %>%
rename(hol.mod.size = value), by = c("holman.module" = "Var1"))
# List all the Morandin et al. modules that significantly overlap with our own modules
overlaps <- data.frame(
overlaps,
do.call("rbind",
lapply(1:nrow(overlaps),
function(i) with(overlaps,
overlap.hypergeometric.test(
overlaps[i],
mor.mod.size[i],
hol.mod.size[i],
nrow(morandin.oggs),
species = "xx"))))[,2:3]) %>%
filter(Test == "Overlap is higher than expected:") %>%
arrange(p) %>% select(-Test) %>%
left_join(morandin.oggs %>%
select(morandin.module, caste.bias) %>%
distinct(), by = "morandin.module") %>%
mutate(p = p.adjust(p, method = "BH"),
morandin.module = paste("Module", morandin.module)) %>%
filter(p < 0.05)
names(overlaps) <- c("Morandin module", "Holman module", "n overlapping genes", "Size of Morandin module", "Size of Holman module", "p-value", "Caste bias of Morandin module")Table S17: A list of the six module pairs, from Morandin et al. 2016 and the present study, which had significantly more genes in common than expected by chance. The p-values were calculated by running hypergeometric tests on all possible pairs of modules from the two studies, and then adjusting all the p-values using the Benjamini-Hochberg procedure.
saveRDS(overlaps, file = "supplement/tab_S17.rds")
overlaps %>% pander(split.cell = 40, split.table = Inf)| Morandin module | Holman module | n overlapping genes | Size of Morandin module | Size of Holman module | p-value | Caste bias of Morandin module |
|---|---|---|---|---|---|---|
| Module 32 | Module 2 | 20 | 39 | 363 | 8.886e-09 | Worker-biased |
| Module 31 | Module 1 | 69 | 161 | 969 | 0.0002602 | Queen-biased |
| Module 26 | Module 8 | 5 | 49 | 24 | 0.0005457 | Worker-biased |
| Module 32 | Module 8 | 4 | 39 | 24 | 0.003328 | Worker-biased |
| Module 13 | Module 4 | 10 | 61 | 177 | 0.01554 | Queen-biased |
| Module 10 | Module 3 | 10 | 77 | 150 | 0.02318 | Worker-biased |
A list of the genes (OGGs) that appear in the present study’s Module 1 (which is pheromone-sensitive), and also in Morandin et al.’s Module 31 (which is caste-biased).
morandin.oggs %>%
filter(morandin.module == 31, holman.module == "Module 1") %>%
select(am, apis.name) %>% rename(`Apis ortholog ID` = am, `Apis ortholog name` = apis.name) %>%
kable.table()| Apis ortholog ID | Apis ortholog name |
|---|---|
| GB54359 | uncharacterized protein LOC726251 isoform X1 |
| GB41231 | ubiquitin-conjugating enzyme E2 G1 |
| GB51699 | WD repeat-containing protein 43-like |
| GB53328 | DNA/RNA-binding protein KIN17 |
| GB42693 | la protein homolog |
| GB47191 | uncharacterized protein LOC100576348 isoform X2 |
| GB50004 | protein preli-like isoform 1 |
| GB50996 | DNA-directed RNA polymerase II subunit RPB3-like |
| GB40765 | ankyrin repeat domain-containing protein 12-like isoform X3 |
| GB42664 | probable ATP-dependent RNA helicase DDX43-like |
| GB42525 | zinc finger protein ZPR1 |
| GB53005 | gem-associated protein 8-like |
| GB41769 | actin-related protein 8 isoform 1 |
| GB45492 | protein misato-like isoform X1 |
| GB54462 | protein LLP homolog |
| GB46707 | vacuolar protein-sorting-associated protein 25 |
| GB51900 | surfeit locus protein 6 homolog |
| GB49621 | alpha-L-fucosidase |
| GB53359 | alkylated DNA repair protein alkB homolog 1 |
| GB40269 | nucleolar protein 14 homolog isoform X1 |
| GB40308 | hsp70-binding protein 1-like |
| GB50555 | thioredoxin peroxidase 3 isoform 2 |
| GB49654 | zinc finger protein 830-like |
| GB41153 | peroxisomal membrane protein PEX16-like |
| GB53829 | CD2 antigen cytoplasmic tail-binding protein 2 homolog |
| GB42223 | elongation factor G, mitochondrial-like |
| GB46495 | tetratricopeptide repeat protein 4 |
| 724533 | small nuclear ribonucleoprotein Sm D3 isoform X1 |
| GB53716 | estrogen sulfotransferase-like |
| GB46074 | survival motor neuron protein-like |
| GB40415 | protein brambleberry-like |
| GB42058 | helicase SKI2W |
| GB47633 | serine/threonine-protein kinase D3 isoformX2 |
| GB40949 | homologous-pairing protein 2 homolog |
| GB44449 | putative 28S ribosomal protein S5, mitochondrial isoform X1 |
| GB45452 | AP-3 complex subunit mu-1-like isoform X1 |
| GB54526 | mitotic spindle assembly checkpoint protein MAD1 |
| GB42778 | U6 snRNA-associated Sm-like protein LSm4-like |
| GB46398 | thyrotroph embryonic factor isoformX1 |
| GB43200 | tRNA (uracil-5-)-methyltransferase homolog A-like isoform X2 |
| GB43625 | RNA methyltransferase-like protein 1-like |
| GB46881 | COMM domain-containing protein 5-like isoform X2 |
| GB54300 | probable U2 small nuclear ribonucleoprotein A’ isoform X1 |
| GB50740 | ubiquitin-conjugating enzyme E2 S-like isoform X2 |
| GB51106 | mRNA turnover protein 4 homolog |
| GB40833 | UPF0396 protein CG6066-like isoform X3 |
| GB41176 | polyglutamine-binding protein 1-like |
| GB10936 | U4/U6 small nuclear ribonucleoprotein Prp3 |
| GB50231 | protein RMD5 homolog A-like |
| GB50167 | probable 28S ribosomal protein S26, mitochondrial |
| 102656337 | mitotic spindle assembly checkpoint protein MAD2A-like isoform X1 |
| GB53707 | density-regulated protein-like isoform X1 |
| GB46986 | 39S ribosomal protein L46, mitochondrial |
| GB53934 | rho GTPase-activating protein 24-like |
| GB44172 | protein CASC3-like isoform X2 |
| GB46452 | DNA replication factor Cdt1 isoform X1 |
| GB53960 | spliceosome-associated protein CWC15 homolog isoform 1 |
| GB41603 | PTB domain-containing adapter protein ced-6 isoform X2 |
| GB50886 | mediator of RNA polymerase II transcription subunit 30 isoform 1 |
| GB47965 | MKI67 FHA domain-interacting nucleolar phosphoprotein-like |
| GB46654 | ribosomal L1 domain-containing protein CG13096-like |
| GB42204 | rho guanine nucleotide exchange factor 3-like |
| GB54976 | ER membrane protein complex subunit 8/9 homolog |
| GB50197 | splicing factor 3A subunit 3 isoform X1 |
| GB46606 | peptidyl-prolyl cis-trans isomerase FKBP8-like isoform X3 |
| GB40355 | coiled-coil domain-containing protein 94-like isoform X2 |
| GB49556 | DNA replication licensing factor Mcm2-like isoform X1 |
| GB51427 | bromodomain-containing protein DDB_G0280777 |
| GB52645 | pinin |
A list of the genes (OGGs) that appear in the present study’s Module 4 (which is pheromone-sensitive), and also in Morandin et al.’s Module 13 (which is caste-biased).
morandin.oggs %>%
filter(morandin.module == 13, holman.module == "Module 4") %>%
select(am, apis.name) %>% rename(`Apis ortholog ID` = am, `Apis ortholog name` = apis.name) %>%
pander(split.cell = 40, split.table = Inf)| Apis ortholog ID | Apis ortholog name |
|---|---|
| GB54848 | tumor suppressor candidate 3-like |
| GB53661 | methyltransferase-like isoform X3 |
| GB50524 | uncharacterized protein LOC726417 |
| GB54153 | uncharacterized protein LOC100576236 isoform X1 |
| GB54999 | NAD kinase 2, mitochondrial-like |
| GB47507 | histone H2A-like |
| GB42799 | protein takeout-like |
| GB42899 | uncharacterized protein LOC551133 isoform X2 |
| GB43984 | xenotropic and polytropic retrovirus receptor 1 homolog |
| GB43942 | putative serine protease K12H4.7-like isoform X2 |
GO and KEGG enrichment for differentially expressed genes
First, define some functions we will need. See also the R script Script to set up for GO analyses.R, which was used to get the GO and KEGG data for Apis.
library(GO.db)
library(AnnotationHub)
select <- dplyr::select
rename <- dplyr::rename
filter <- dplyr::filter
if(!file.exists("data/component spreadsheets of queen_pheromone.db/all_apis_go_terms.rds")){
library(AnnotationHub)
# Connect to AnnotationHub, and get the annotations for A mellifera
hub <- AnnotationHub::AnnotationHub()
apis.db <- hub[["AH62534"]]
apis_go_table <- dbconn(apis.db) %>% tbl("go") %>% collect() # Connect to GO table in the database
apis_go <- tbl(dbconn(apis.db), "go") %>% left_join(tbl(dbconn(apis.db), "entrez_genes")) %>%
select(ENTREZID, GO, ONTOLOGY) %>%
rename(gene = ENTREZID, ontology = ONTOLOGY) %>% collect(n = Inf)
apis_entrez_names <- tbl(dbconn(apis.db), "gene_info") %>% left_join(tbl(dbconn(apis.db), "entrez_genes")) %>%
select(ENTREZID, GENENAME) %>%
rename(entrez = ENTREZID, name = GENENAME) %>% collect(n = Inf)
go_meanings <- dbconn(GO.db) %>% tbl("go_term") %>% select(go_id, term, ontology) %>% collect()
saveRDS(apis.go, file = "data/component spreadsheets of queen_pheromone.db/all_apis_go_terms.rds")
saveRDS(go_meanings, file = "data/component spreadsheets of queen_pheromone.db/go_meanings.rds")
my_db$con %>% db_drop_table(table = "apis_go")
copy_to(my_db, apis_go, "apis_go", temporary = FALSE)
my_db$con %>% db_drop_table(table = "apis_entrez_names")
copy_to(my_db, apis_entrez_names, "apis_entrez_names", temporary = FALSE)
my_db$con %>% db_drop_table(table = "go_meanings")
copy_to(my_db, go_meanings, "go_meanings", temporary = FALSE)
}
# Makes a database shortcut (lazy query) to a table of parent and child GO term relationships, from the massive GO.db database
make_db_shortcut <- function(tabl){
dbconn(GO.db) %>% tbl("go_bp_offspring") %>%
left_join(dbconn(GO.db) %>% tbl("go_term") %>% select(`_id`, go_id), by = "_id") %>% rename(parent_GO = go_id) %>%
left_join(dbconn(GO.db) %>% tbl("go_term") %>% select(`_id`, go_id), by = c("_offspring_id" = "_id")) %>%
rename(child_GO = go_id) %>% select(parent_GO, child_GO)
}
# Function to get all the descendants of a focal GO term, given its "hierarchy" (i.e. one of the lazy db queries BP, MF, or CC)
get_descendants <- function(ancestor, hierarchy) hierarchy %>% filter(parent_GO == ancestor) %>% select(child_GO) %>% collect() %>% .$child_GO
# Fucntion to run GSEA implemented in the fgsea package
GO.and.KEGG.gsea <- function(tabl = NULL, df = NULL, min.size = 5, keep.all = FALSE){
p <- 0.05; if(keep.all) p <- 1 # Set the significance threshold
if(!is.null(tabl)){
# First, grab the data for the focal species. If not Apis, then get the names of the Apis orthologs via reciprocal best BLAST
if(tabl == "ebseq_gene_am") {df <- tbl(my_db, tabl) %>% collect()}
if(tabl == "ebseq_gene_bt") {
df <- tbl(my_db, "ebseq_gene_bt") %>% collect() %>%
left_join(make.OGGs(c("am", "bt"))[[2]], by = c("gene" = "bt")) %>%
dplyr::select(-gene) %>% rename(gene = am) %>% filter(!is.na(gene))
}
if(tabl == "ebseq_gene_lf") {
df <- tbl(my_db, "ebseq_gene_lf") %>% collect() %>%
left_join(make.OGGs(c("am", "lf"))[[2]], by = c("gene" = "lf")) %>%
select(-gene) %>% rename(gene = am) %>% filter(!is.na(gene))
}
if(tabl == "ebseq_gene_ln") {
df <- tbl(my_db, "ebseq_gene_ln") %>% collect() %>%
left_join(make.OGGs(c("am", "ln"))[[2]], by = c("gene" = "ln")) %>%
select(-gene) %>% rename(gene = am) %>% filter(!is.na(gene))
}
# Calculate pheromone sensitivity for each gene
df <- df %>% mutate(sensitivity = abs(log2(PostFC)) ) %>% arrange(-sensitivity) %>% as.data.frame()
}
# Set up the geneList object in the form needed by the fgsea function - named, ranked vector of pheromone sensitivity per gene
geneList <- df$sensitivity
names(geneList) <- entrez.tbl$entrez.id[match(df$gene, entrez.tbl$gene)] # Convert Beebase2 to Entrez gene names
gene_universe <- names(geneList)
# Internal function to run GO enrichment
GO.enrichment <- function(geneList, ontol){
pathways <- tbl(my_db, "apis_go") %>%
filter(gene %in% gene_universe,
ontology == ontol) %>%
select(-ontology) %>% collect(n=Inf)
pathways <- with(pathways, split(gene, GO))
result <- fgsea::fgsea(pathways, geneList, nperm = 10000, minSize = min.size, maxSize = 500) %>%
filter(pval <= p)
collapse_pathways <- fgsea::collapsePathways(result, pathways, geneList)
pathways_to_keep <- c(collapse_pathways[[1]], names(collapse_pathways[[2]]))
result <- result %>% filter(pathway %in% pathways_to_keep)
result <- result %>%
rename(ID = pathway) %>%
left_join(tbl(my_db, "go_meanings") %>%
select(-ontology) %>% collect(n=Inf), by = c("ID" = "go_id"))
if(nrow(result) == 0) return(NULL)
if(ontol == "BP") Test_type <- "GO: Biological process"
if(ontol == "MF") Test_type <- "GO: Molecular function"
if(ontol == "CC") Test_type <- "GO: Cellular component"
data.frame(Test_type = Test_type, result, stringsAsFactors = FALSE)
}
# Internal function to run KEGG enrichment
kegg.enrichment <- function(geneList){
apis_kegg <- clusterProfiler::download_KEGG("ame")
apis_kegg_names <- apis_kegg[[2]]
apis_kegg_focal <- apis_kegg[[1]] %>% filter(to %in% gene_universe)
pathways <- with(apis_kegg_focal, split(to, from))
result <- fgsea::fgsea(pathways, geneList, nperm = 10000, minSize = min.size, maxSize = 500) %>%
filter(pval <= p)
collapse_pathways <- fgsea::collapsePathways(result, pathways, geneList)
result <- result %>% filter(pathway %in% c(collapse_pathways[[1]], names(collapse_pathways[[2]]))) %>%
rename(ID = pathway) %>%
left_join(apis_kegg_names %>% rename(term = to), by = c("ID" = "from")) %>%
mutate(ID = str_replace_all(ID, "ame", "KEGG:"))
if(nrow(result) == 0) return(NULL)
data.frame(Test_type = "KEGG", result, stringsAsFactors = FALSE)
}
rbind(GO.enrichment(geneList, "BP"),
GO.enrichment(geneList, "MF"),
GO.enrichment(geneList, "CC"),
kegg.enrichment(geneList))
}
# The previous function has a call to collapse_pathways(), which is great for finding over-arching patterns, but it results in a patchy version of Figure 2. This is because the pathways get collapsed slightly differently in each species, based on which genes happen to have detectable orthologs in A. mellifera, plus the gene expression data itself. This function attempts to fill all the holes in Figure 2 by running additional GSEA tests on all the GO terms that are missing from the figure. A small number of holes remain in Figure 2 due to missing orthologs in the non-Apis species, but mostly it does a good job.
test_child_pathways <- function(row, job_list, three_hierarchies, ebseq_tables, apis_go, go_meanings, three_OGGs, entrez.tbl){ # input: a dataframe of jobs, and the row of the job that needs doing
# Get the focal GO_id, ontology, species, and GO tree
go_id <- job_list$GO[row]
ontol <- job_list$Test_type[row]
Species <- job_list$Species[row]
if(grepl("Biological", ontol)) {hierarchy <- three_hierarchies[[1]]; short_ontol <- "BP"}
if(grepl("Molecular", ontol)) {hierarchy <- three_hierarchies[[2]]; short_ontol <- "MF"}
if(grepl("Cellular", ontol)) {hierarchy <- three_hierarchies[[3]]; short_ontol <- "CC"}
# Get the relevant data needed to rank the genes and make the 'geneList' object
if(Species == "am") df <- ebseq_tables[[1]]
if(Species == "bt") {
df <- ebseq_tables[[2]] %>%
left_join(three_OGGs[[1]], by = c("gene" = "bt")) %>%
dplyr::select(-gene) %>% rename(gene = am) %>% filter(!is.na(gene))
}
if(Species == "lf") {
df <- ebseq_tables[[3]] %>%
left_join(three_OGGs[[2]], by = c("gene" = "lf")) %>%
dplyr::select(-gene) %>% rename(gene = am) %>% filter(!is.na(gene))
}
if(Species == "ln") {
df <- ebseq_tables[[4]] %>%
left_join(three_OGGs[[3]], by = c("gene" = "ln")) %>%
dplyr::select(-gene) %>% rename(gene = am) %>% filter(!is.na(gene))
}
# Set up the geneList object in the form needed by the fgsea function - named, ranked vector of pheromone sensitivity per gene
df <- df %>% mutate(sensitivity = abs(log2(PostFC))) %>% arrange(-sensitivity) %>% as.data.frame()
geneList <- df$sensitivity
names(geneList) <- entrez.tbl$entrez.id[match(df$gene, entrez.tbl$gene)] # Convert Beebase2 to Entrez gene names
gene_universe <- names(geneList)
# Run the GSEA, just on the focal GO term (and its descendants)
pathways <- apis_go %>% filter(gene %in% gene_universe,
ontology == short_ontol) %>% select(-ontology)
pathways$GO[pathways$GO %in% get_descendants(go_id, hierarchy)] <- go_id
pathways <- with(pathways, split(gene, GO))
foc <- which(names(pathways) == go_id)
result <- fgsea::fgsea(pathways[foc], geneList, nperm = 10000)
if(nrow(result) > 0) {
result <- rename(result, ID = pathway) %>%
left_join(go_meanings %>% select(-ontology), by = c("ID" = "go_id"))
return(data.frame(Test_type = ontol, result, Species, stringsAsFactors = FALSE))
}
else return(NULL)
}
# Function that attempts to fill in the gaps using test_child_pathways()
fill_in_GSEA_results <- function(GSEA_results){
# Remove all KEGG and GO terms except those that are significant in at least one species
results <- GSEA_results %>%
filter(!is.nan(NES)) %>%
filter(ID %in% (GSEA_results %>% filter(pval < 0.05) %>% .$ID %>% unique))
# List GO terms that were not measured in all 4 species (often because of the GO concatenation step above)
incomplete_GOs <- results %>%
filter(Test_type != "KEGG") %>%
group_by(Test_type, ID) %>% summarise(n = n()) %>% filter(!(n %in% c(4, 8, 12))) %>% .$ID
job_list <- expand.grid(Species = c("am", "bt", "lf", "ln"),
GO = incomplete_GOs) %>%
filter(!(paste(Species, GO) %in% paste(results$Species, results$GO))) %>%
left_join(results %>% rename(GO = ID) %>% select(GO, Test_type) %>% distinct(), by = "GO")
# Remove combinations that were already run from the job_list
job_list <- job_list %>% filter(!(paste(Species, GO) %in% paste(GSEA_results$Species, GSEA_results$ID)))
# Pull all of this information into memory, so that we can use mclapply for speed
# (mclapply does not play well with database connections)
three_hierarchies <- list(
make_db_shortcut("go_bp_offspring") %>% collect(),
make_db_shortcut("go_mf_offspring") %>% collect(),
make_db_shortcut("go_cc_offspring") %>% collect()
)
ebseq_tables <- list(
tbl(my_db, "ebseq_gene_am") %>% collect(),
tbl(my_db, "ebseq_gene_bt") %>% collect(),
tbl(my_db, "ebseq_gene_lf") %>% collect(),
tbl(my_db, "ebseq_gene_ln") %>% collect()
)
apis_go <- tbl(my_db, "apis_go") %>% collect()
go_meanings <- tbl(my_db, "go_meanings") %>% collect()
three_OGGs <- list(
make.OGGs(c("am", "bt"))[[2]],
make.OGGs(c("am", "lf"))[[2]],
make.OGGs(c("am", "ln"))[[2]]
)
rbind(results,
lapply(1:nrow(job_list), function(i)
test_child_pathways(i, job_list, three_hierarchies, ebseq_tables, apis_go, go_meanings, three_OGGs, entrez.tbl)) %>% do.call("rbind", .)) %>%
distinct() %>%
rename(Description = term, pvalue = pval, p.adjust = padj) %>%
filter(!is.nan(NES)) %>%
mutate(p.adjust = p.adjust(pvalue, method = "BH")) %>%
arrange(Test_type, Species, pvalue) %>% ungroup()
}
# Run the GO and KEGG enrichment analyses using GSEA implemented in fgsea
# The functions here run GSEA on each species and all 4 ontologies,
# then collapse redundant, smaller GO terms into higher-order ones, and then fill in any gaps to make Figure 2 less patchy
set.seed(1)
GO_and_KEGG_results_GSEA <- rbind(
GO.and.KEGG.gsea("ebseq_gene_am", keep.all = TRUE) %>% mutate(Species = "am"),
GO.and.KEGG.gsea("ebseq_gene_bt", keep.all = TRUE) %>% mutate(Species = "bt"),
GO.and.KEGG.gsea("ebseq_gene_lf", keep.all = TRUE) %>% mutate(Species = "lf"),
GO.and.KEGG.gsea("ebseq_gene_ln", keep.all = TRUE) %>% mutate(Species = "ln")) %>%
fill_in_GSEA_results() GSEA for pheromone sensitivity of gene expression
Figure 2 provides a summary of the significant and/or interesting enriched GO terms, and is intended to provide a compact summary of the next 3 figures, which show every GO term for which we had data on at least 5 genes.
make_enrichment_heatmap <- function(GO_and_KEGG_results, pval_cutoff){
plot_data <- GO_and_KEGG_results %>%
mutate(sig = " ",
sig = replace(sig, pvalue < 0.05, "*"),
sig = replace(sig, p.adjust < 0.05, "**"))
levels <- plot_data %>%
group_by(Test_type, Description) %>%
summarise(summed_NES = sum(NES), n = n()) %>%
arrange(desc(Test_type), summed_NES) %>% .$Description
grid_arrange_shared_legend <- function(p1, p2) {
plots <- list(p1, p2)
g <- ggplotGrob(plots[[1]] + theme(legend.position="right"))$grobs
legend <- g[[which(sapply(g, function(x) x$name) == "guide-box")]]
lwidth <- sum(legend$width)
arrangeGrob(
arrangeGrob(p1 + theme(legend.position="none"),
p2 + theme(legend.position="none"), ncol = 2, widths = c(0.56,0.44)),
legend,
ncol = 2,
widths = unit.c(unit(1, "npc") - lwidth, lwidth))
}
output <- grid_arrange_shared_legend(
plot_data %>% mutate(Description = factor(Description, levels)) %>%
filter(Test_type %in% c("GO: Biological process", "GO: Molecular function")) %>%
ggplot(aes(Species, Description, fill = NES)) +
geom_tile(colour = "grey10", size = 0.4, linetype = 3) +
geom_text(aes(label = sig)) +
scale_fill_distiller(palette = "RdYlBu") +
scale_y_discrete(labels = function(x) str_wrap(x, width = 55), expand = c(0,0)) +
scale_x_discrete(expand = c(0,0)) +
facet_grid(rows = vars(Test_type), scales = "free_y", space = "free_y") +
theme_minimal() +
theme(panel.border = element_rect(size = 0.8, colour = "grey10", fill = NA),
strip.text = element_text(size = 10)) +
guides(fill = guide_colourbar(frame.colour = "grey10", ticks.colour = "grey10")) +
ylab(NULL) + xlab(NULL),
plot_data %>% mutate(Description = factor(Description, levels)) %>%
filter(Test_type %in% c("GO: Cellular component", "KEGG")) %>%
ggplot(aes(Species, Description, fill = NES)) +
geom_tile(colour = "grey10", size = 0.4, linetype = 3) +
geom_text(aes(label = sig)) +
scale_fill_distiller(palette = "RdYlBu") +
scale_y_discrete(labels = function(x) str_wrap(x, width = 30), expand = c(0,0)) +
scale_x_discrete(expand = c(0,0)) +
facet_grid(rows = vars(Test_type), scales = "free_y", space = "free_y") +
theme_minimal() +
theme(panel.border = element_rect(size = 0.8, colour = "grey10", fill = NA),
strip.text = element_text(size = 10)) +
guides(fill = guide_colourbar(frame.colour = "grey10", ticks.colour = "grey10")) +
ylab(NULL) + xlab(NULL))
grobTree(rectGrob(gp = gpar(fill="white", lwd = 0)), output)
}
figure2 <- make_enrichment_heatmap(GO_and_KEGG_results_GSEA, pval_cutoff = 0.05)
ggsave(figure2, file = "figures/Figure 2 - GO and KEGG.pdf", height = 11.8, width = 9.5)
grid.draw(figure2)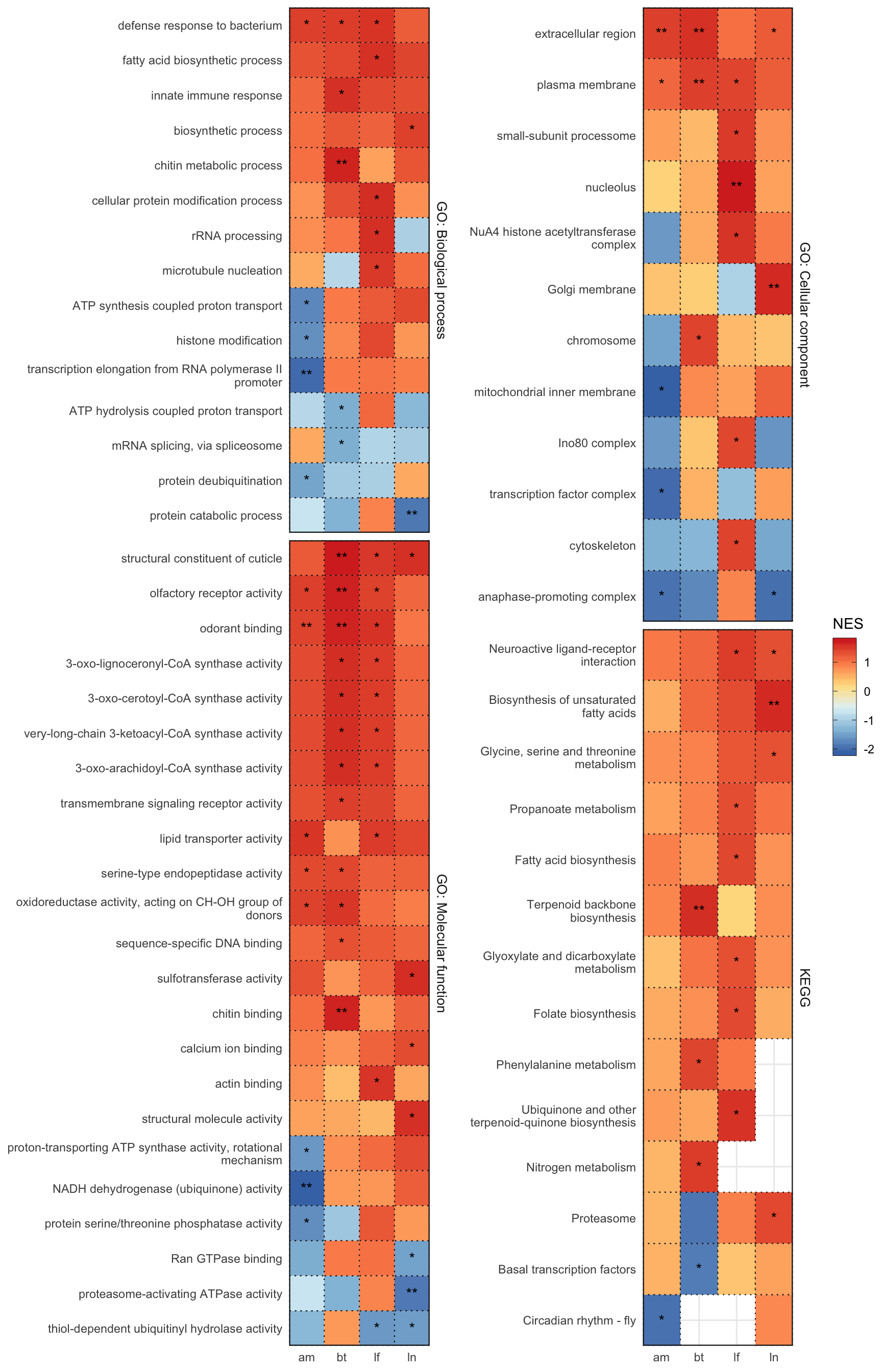
Figure 2: A list of all the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms that were significantly enriched or de-enriched among pheromone-sensitive genes in at least one of the four species. The colour shows the normalised expression score from gene set enrichment analysis; positive (red) values indicate that the ontology term is over-represented among genes whose expression is strongly affected by queen pheromone, and negative (blue) values indicate under-representation among these genes. Asterisks denote statistically significant enrichment (p < 0.05), and double asterisks mark results that remained significant after Benjamini-Hochberg correction. Empty squares denote cases where we did not find at least 5 genes annotated with the focal term.
Tabular results for GSEA for pheromone sensitivity in gene expression
Click the tabs to see tables for each of the four species.
Apis mellifera
Table S18: The results of GSEA (gene set enrichment analysis) for pheromone sensitivity in gene expression in Apis mellifera. The table lists GO and KEGG terms with their NES (normalized enrichment score), the associated raw and adjusted p-values (adjustment was performed using Benjamini-Hochberg correction), and the genes underlying the enrichment result.
add_name_col <- function(df){
sapply(df$leadingEdge, function(x){
data.frame(gene = x, stringsAsFactors = FALSE) %>%
left_join(tbl(my_db, "apis_entrez_names") %>% collect(), by = c("gene" = "entrez")) %>% .$name %>%
paste0(collapse = "; ")
}) -> df$gene_names
df %>% dplyr::rename(enriched_gene_ids = leadingEdge) %>%
mutate(enriched_gene_ids = map_chr(enriched_gene_ids, function(x) paste0(x, collapse = " ")))
}
tab_S18 <- GO_and_KEGG_results_GSEA %>%
filter(Species == "am" & pvalue < 0.05) %>%
arrange(Test_type, pvalue) %>%
select(-Species, -ES, -nMoreExtreme, -size) %>%
add_name_col()
saveRDS(tab_S18 %>% select(-enriched_gene_ids, -gene_names), file = "supplement/tab_S18.rds")
kable.table(tab_S18)| Test_type | ID | pvalue | p.adjust | NES | enriched_gene_ids | Description | gene_names |
|---|---|---|---|---|---|---|---|
| GO: Biological process | GO:0006368 | 0.0006892 | 0.0322997 | -2.019849 | 551906 409928 412377 725696 552621 | transcription elongation from RNA polymerase II promoter | another transcription unit protein; RNA polymerase II elongation factor Ell; parafibromin; RNA polymerase-associated protein Rtf1; RNA polymerase II-associated factor 1 homolog |
| GO: Biological process | GO:0015986 | 0.0064935 | 0.0773036 | -1.737779 | 551766 409114 552682 409148 409236 551861 726120 727483 552699 | ATP synthesis coupled proton transport | ATP synthase subunit beta, mitochondrial; ATP synthase subunit alpha, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9); ATP synthase subunit O, mitochondrial; ATP synthase subunit e, mitochondrial; ATP synthase-coupling factor 6, mitochondrial; ATP synthase subunit epsilon, mitochondrial-like; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| GO: Biological process | GO:0016570 | 0.0165403 | 0.0984543 | -1.652894 | 551906 412377 725696 552621 413130 | histone modification | another transcription unit protein; parafibromin; RNA polymerase-associated protein Rtf1; RNA polymerase II-associated factor 1 homolog; RNA polymerase-associated protein CTR9 homolog |
| GO: Biological process | GO:0042742 | 0.0327906 | 0.1343292 | 1.461421 | 406143 406142 725074 406140 | defense response to bacterium | defensin 1; hymenoptaecin; omega-conotoxin-like protein 1; apidaecin 1 |
| GO: Biological process | GO:0016579 | 0.0447761 | 0.1393728 | -1.457013 | 413813 411920 412114 409389 410109 408619 724338 410344 411572 410162 411362 552324 724537 552660 409403 408911 725744 412644 411981 | protein deubiquitination | ubiquitin carboxyl-terminal hydrolase isozyme L5; ubiquitin carboxyl-terminal hydrolase 22; ubiquitin carboxyl-terminal hydrolase 35-like; ubiquitin specific protease-like; ubiquitin carboxyl-terminal hydrolase 34; ubiquitin carboxyl-terminal hydrolase 3-like; OTU domain-containing protein 5-B; ubiquitin carboxyl-terminal hydrolase CYLD; ubiquitin carboxyl-terminal hydrolase 14; ataxin-3-like; ubiquitin carboxyl-terminal hydrolase 8-like; ubiquitin carboxyl-terminal hydrolase 5; ubiquitin carboxyl-terminal hydrolase 36; ubiquitin carboxyl-terminal hydrolase 46; ubiquitin carboxyl-terminal hydrolase 30 homolog; ubiquitin carboxyl-terminal hydrolase 47; ubiquitin thioesterase otubain-like; josephin-2; probable ubiquitin carboxyl-terminal hydrolase FAF-X |
| GO: Cellular component | GO:0005576 | 0.0001000 | 0.0125000 | 1.535354 | 406121 100577331 406090 406133 406091 406088 410884 406143 406116 406095 503862 100049551 725725 724880 551268 406140 727193 410337 408365 410751 724246 412887 413705 406083 551407 725217 410928 411830 | extracellular region | major royal jelly protein 3; cell wall integrity and stress response component 1-like; major royal jelly protein 1; major royal jelly protein 4; major royal jelly protein 2; vitellogenin; phospholipase A1; defensin 1; major royal jelly protein 5; serine protease 34; corazonin; bursicon subunit alpha; peritrophin-1-like; allatostatin A; pancreatic triacylglycerol lipase-like; apidaecin 1; lipase member H-A-like; venom dipeptidylpeptidase IV; uncharacterized LOC408365; phospholipase A1 member A; growth/differentiation factor 2-like; A disintegrin and metalloproteinase with thrombospondin motifs 7-like; acidic mammalian chitinase-like; tachykinin; A disintegrin and metalloproteinase with thrombospondin motifs 14-like; uncharacterized protein PFB0145c-like; venom carboxylesterase-6; venom acid phosphatase |
| GO: Cellular component | GO:0005667 | 0.0363014 | 0.1343292 | -1.536247 | 409321 409887 410757 409301 412770 | transcription factor complex | mothers against decapentaplegic homolog 4; transcription factor Dp-1; circadian locomoter output cycles protein kaput; protein mothers against dpp; transcription factor E2F2 |
| GO: Cellular component | GO:0005743 | 0.0384615 | 0.1343292 | -1.636964 | 406075 408968 413014 725881 727493 551169 726316 724264 550667 551811 726314 552482 724499 100577081 725566 408734 408548 725527 551337 413781 411790 411677 408837 412409 413186 | mitochondrial inner membrane | ADP/ATP translocase; cytochrome c-type heme lyase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; uncharacterized LOC725881; ubiquinone biosynthesis protein COQ4 homolog, mitochondrial-like; succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial-like; cytochrome b-c1 complex subunit 6, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial; graves disease carrier protein homolog; protein SCO2 homolog, mitochondrial; protoporphyrinogen oxidase; mitochondrial pyruvate carrier 2-like; dihydroorotate dehydrogenase (quinone), mitochondrial; succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial; succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial-like; MICOS complex subunit Mic60; cytochrome b-c1 complex subunit Rieske, mitochondrial; flotillin-1; surfeit locus protein 1; solute carrier family 25 member 38-like; mitochondrial pyruvate carrier 1; cytochrome c oxidase subunit 5A, mitochondrial; mitochondrial import inner membrane translocase subunit TIM44; mitochondrial pyruvate carrier 3-like |
| GO: Cellular component | GO:0005680 | 0.0444840 | 0.1393728 | -1.464557 | 410290 726378 413293 409810 413499 724920 | anaphase-promoting complex | anaphase-promoting complex subunit 5; anaphase-promoting complex subunit 11-like; anaphase-promoting complex subunit 10; anaphase-promoting complex subunit 4; cell division cycle protein 23 homolog; anaphase-promoting complex subunit 15-like |
| GO: Cellular component | GO:0005886 | 0.0488951 | 0.1438091 | 1.232225 | 100576816 100578083 725205 412740 100577755 100577334 100577482 725052 552546 100577062 411760 100576167 100577938 100578189 100576681 100578402 100577446 552785 100577590 | plasma membrane | odorant receptor 4-like; odorant receptor 14; odorant receptor 1; ligand-gated chloride channel homolog 3; odorant receptor 30a-like; odorant receptor 33; odorant receptor 25; odorant receptor 13a; protocadherin Fat 4; odorant receptor 4-like; metabotropic glutamate receptor 7; gustatory and pheromone receptor 32a-like; odorant receptor 18; odorant receptor 63; odorant receptor 57; odorant receptor 4; odorant receptor 27; ligand-gated ion channel pHCl; odorant receptor 20 |
| GO: Molecular function | GO:0008137 | 0.0011628 | 0.0322997 | -2.129582 | 411411 725881 408909 724827 551660 725315 724264 | NADH dehydrogenase (ubiquinone) activity | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial; uncharacterized LOC725881; NADH-quinone oxidoreductase subunit B 2; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial; uncharacterized LOC725315; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| GO: Molecular function | GO:0005549 | 0.0021998 | 0.0419353 | 1.432831 | 677675 100576816 100578083 725205 677673 100577755 100577334 100577482 725052 551719 100577062 100577938 100578189 100576681 406109 100578402 100577446 406101 100577590 410068 | odorant binding | odorant binding protein 9; odorant receptor 4-like; odorant receptor 14; odorant receptor 1; odorant binding protein 14; odorant receptor 30a-like; odorant receptor 33; odorant receptor 25; odorant receptor 13a; odorant binding protein 16; odorant receptor 4-like; odorant receptor 18; odorant receptor 63; odorant receptor 57; odorant binding protein 6; odorant receptor 4; odorant receptor 27; odorant binding protein 4; odorant receptor 20; odorant binding protein 10 |
| GO: Molecular function | GO:0004984 | 0.0050010 | 0.0694583 | 1.460117 | 100576816 100578083 725205 100577755 100577334 100577482 725052 100577062 100577938 100578189 100576681 100578402 100577446 100577590 | olfactory receptor activity | odorant receptor 4-like; odorant receptor 14; odorant receptor 1; odorant receptor 30a-like; odorant receptor 33; odorant receptor 25; odorant receptor 13a; odorant receptor 4-like; odorant receptor 18; odorant receptor 63; odorant receptor 57; odorant receptor 4; odorant receptor 27; odorant receptor 20 |
| GO: Molecular function | GO:0004722 | 0.0104651 | 0.0872093 | -1.667797 | 408701 409804 409430 551020 552412 552068 413080 | protein serine/threonine phosphatase activity | protein phosphatase 1H; serine/threonine-protein phosphatase PP1-beta catalytic subunit; serine/threonine-protein phosphatase alpha-2 isoform; probable protein phosphatase 2C T23F11.1; probable protein phosphatase CG10417; protein phosphatase 1L; pyruvate dehydrogenase [acetyl-transferring]-phosphatase 1, mitochondrial |
| GO: Molecular function | GO:0005319 | 0.0124725 | 0.0922074 | 1.537033 | 406088 726182 408992 | lipid transporter activity | vitellogenin; larval-specific very high density lipoprotein; Niemann-Pick C1 protein-like |
| GO: Molecular function | GO:0004252 | 0.0199040 | 0.1130908 | 1.364744 | 724565 724477 406095 409204 410894 409827 409143 413645 408534 724308 100576326 411358 724208 | serine-type endopeptidase activity | trypsin-7; vitamin K-dependent protein C; serine protease 34; trypsin; chymotrypsin-1; serine proteinase stubble; venom serine protease 34; trypsin alpha-3; trypsin; trypsin-1; trypsin-7; trypsin-1; venom protease-like |
| GO: Molecular function | GO:0046933 | 0.0232558 | 0.1211240 | -1.588367 | 551766 409114 552682 725661 409236 727483 552699 | proton-transporting ATP synthase activity, rotational mechanism | ATP synthase subunit beta, mitochondrial; ATP synthase subunit alpha, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; uncharacterized LOC725661; ATP synthase subunit O, mitochondrial; ATP synthase subunit epsilon, mitochondrial-like; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| GO: Molecular function | GO:0016614 | 0.0271038 | 0.1254808 | 1.440299 | 727367 552457 410746 410744 410748 551044 552395 | oxidoreductase activity, acting on CH-OH group of donors | glucose dehydrogenase [FAD, quinone]-like; glucose dehydrogenase [FAD, quinone]; glucose dehydrogenase [FAD, quinone]; glucose dehydrogenase [FAD, quinone]; glucose dehydrogenase [FAD, quinone]; glucose dehydrogenase [FAD, quinone]; glucose dehydrogenase [FAD, quinone] |
| KEGG | KEGG:04711 | 0.0373726 | 0.1343292 | -1.477501 | 410253 725614 408449 412108 410757 406112 408976 | Circadian rhythm - fly | ataxin-2 homolog; protein cycle; thyrotroph embryonic factor; casein kinase I-like; circadian locomoter output cycles protein kaput; period circadian protein; protein kinase shaggy |
Bombus terrestris
Table S19: The results of GSEA (gene set enrichment analysis) for pheromone sensitivity in gene expression in Bombus terrestris. The table lists GO and KEGG terms with their NES (normalized enrichment score), the associated raw and adjusted p-values (adjustment was performed using Benjamini-Hochberg correction), and the genes underlying the enrichment result.
tab_S19 <- GO_and_KEGG_results_GSEA %>%
filter(Species == "bt" & pvalue < 0.05) %>%
arrange(Test_type, pvalue) %>%
select(-Species, -ES, -nMoreExtreme, -size) %>%
add_name_col()
saveRDS(tab_S19 %>% select(-enriched_gene_ids, -gene_names), file = "supplement/tab_S19.rds")
kable.table(tab_S19)| Test_type | ID | pvalue | p.adjust | NES | enriched_gene_ids | Description | gene_names |
|---|---|---|---|---|---|---|---|
| GO: Biological process | GO:0006030 | 0.0007056 | 0.0322997 | 1.683935 | 551323 724464 413481 413679 725932 408365 100577576 724382 100576184 727129 411273 413560 100577513 | chitin metabolic process | uncharacterized LOC551323; cuticular protein; probable chitinase 3; cuticular protein analogous to peritrophins 3-E; cuticular protein analogous to peritrophins 3-C; uncharacterized LOC408365; chondroitin proteoglycan-2-like; cuticular protein analogous to peritrophins 3-A; uncharacterized LOC100576184; uncharacterized LOC727129; uncharacterized protein DDB_G0287625-like; uncharacterized LOC413560; uncharacterized LOC100577513 |
| GO: Biological process | GO:0045087 | 0.0121690 | 0.0922074 | 1.588706 | 406143 406140 100576745 | innate immune response | defensin 1; apidaecin 1; leucine-rich repeat-containing protein 26-like |
| GO: Biological process | GO:0000398 | 0.0364964 | 0.1343292 | -1.381094 | 725352 413842 551620 725401 551974 552155 409347 413548 725142 411122 725947 413523 408606 413963 552529 408632 | mRNA splicing, via spliceosome | U6 snRNA-associated Sm-like protein LSm7; splicing factor U2af 38 kDa subunit; pre-mRNA-processing-splicing factor 8; U6 snRNA-associated Sm-like protein LSm3; thioredoxin-like protein 4A; U6 snRNA-associated Sm-like protein LSm8; U4/U6.U5 tri-snRNP-associated protein 1; intron-binding protein aquarius; U6 snRNA-associated Sm-like protein LSm2; splicing factor 1; U6 snRNA-associated Sm-like protein LSm6; pre-mRNA-processing factor 17; PRP3 pre-mRNA processing factor 3 homolog; splicing factor 3A subunit 3; U6 snRNA-associated Sm-like protein LSm4; spliceosome-associated protein CWC15 homolog |
| GO: Biological process | GO:0015991 | 0.0417827 | 0.1365156 | -1.376223 | 409148 406076 552410 409074 409946 552720 551093 409055 551721 725661 552476 | ATP hydrolysis coupled proton transport | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9); vacuolar H+ ATP synthase 16 kDa proteolipid subunit; V-type proton ATPase subunit e 2-like; V-type proton ATPase 21 kDa proteolipid subunit-like; V-type proton ATPase subunit d; V-type proton ATPase subunit E; V-type proton ATPase catalytic subunit A; V-type proton ATPase subunit H; V-type proton ATPase subunit B; uncharacterized LOC725661; V-type proton ATPase subunit F |
| GO: Biological process | GO:0042742 | 0.0439150 | 0.1393728 | 1.475540 | 406143 406140 100576745 725074 | defense response to bacterium | defensin 1; apidaecin 1; leucine-rich repeat-containing protein 26-like; omega-conotoxin-like protein 1 |
| GO: Cellular component | GO:0005576 | 0.0001000 | 0.0125000 | 1.650311 | 410751 406143 551323 724464 413481 413679 406140 725932 408365 100577576 503862 724382 100576184 100126690 727129 410337 409307 100576512 410884 410451 411273 413560 724563 100577513 | extracellular region | phospholipase A1 member A; defensin 1; uncharacterized LOC551323; cuticular protein; probable chitinase 3; cuticular protein analogous to peritrophins 3-E; apidaecin 1; cuticular protein analogous to peritrophins 3-C; uncharacterized LOC408365; chondroitin proteoglycan-2-like; corazonin; cuticular protein analogous to peritrophins 3-A; uncharacterized LOC100576184; pheromone biosynthesis-activating neuropeptide; uncharacterized LOC727129; venom dipeptidylpeptidase IV; uncharacterized LOC409307; U8-agatoxin-Ao1a-like; phospholipase A1; venom serine carboxypeptidase; uncharacterized protein DDB_G0287625-like; uncharacterized LOC413560; uncharacterized LOC724563; uncharacterized LOC100577513 |
| GO: Cellular component | GO:0005886 | 0.0027027 | 0.0450450 | 1.544910 | 100578210 100577938 100577446 100577755 725297 100578083 100577334 411760 100578230 725205 551782 | plasma membrane | odorant receptor 5; odorant receptor 18; odorant receptor 27; odorant receptor 30a-like; gustatory receptor 10; odorant receptor 14; odorant receptor 33; metabotropic glutamate receptor 7; odorant receptor 115; odorant receptor 1; bestrophin-4-like |
| GO: Cellular component | GO:0005694 | 0.0260593 | 0.1252852 | 1.511039 | 726963 412750 | chromosome | meiotic recombination protein SPO11; DNA topoisomerase 1 |
| GO: Molecular function | GO:0042302 | 0.0003016 | 0.0251357 | 1.737132 | 413115 725509 727392 726995 726451 726950 727197 724777 725300 724556 100577562 724649 724624 | structural constituent of cuticle | cuticular protein 19; cuticular protein 27; endocuticle structural glycoprotein SgAbd-2; endocuticle structural glycoprotein ABD-4; cuticle protein 7; pupal cuticle protein 20; cuticular protein 2; cuticular protein 14; cuticular protein 13; cuticular protein 17; probable LIM domain-containing serine/threonine-protein kinase DDB_G0286997; cuticular protein 21; endocuticle structural glycoprotein SgAbd-1 |
| GO: Molecular function | GO:0004984 | 0.0009223 | 0.0322997 | 1.703267 | 100578210 100577938 100577446 100577755 100578083 100577334 100578230 725205 | olfactory receptor activity | odorant receptor 5; odorant receptor 18; odorant receptor 27; odorant receptor 30a-like; odorant receptor 14; odorant receptor 33; odorant receptor 115; odorant receptor 1 |
| GO: Molecular function | GO:0005549 | 0.0011033 | 0.0322997 | 1.677641 | 100578210 100577938 100577446 100577755 100578083 100577334 100578230 677674 725205 406103 406109 | odorant binding | odorant receptor 5; odorant receptor 18; odorant receptor 27; odorant receptor 30a-like; odorant receptor 14; odorant receptor 33; odorant receptor 115; odorant binding protein 13; odorant receptor 1; odorant binding protein 2; odorant binding protein 6 |
| GO: Molecular function | GO:0008061 | 0.0011076 | 0.0322997 | 1.678200 | 551323 724464 413481 413679 725932 408365 100577576 724382 100576184 727129 411273 413560 100577513 | chitin binding | uncharacterized LOC551323; cuticular protein; probable chitinase 3; cuticular protein analogous to peritrophins 3-E; cuticular protein analogous to peritrophins 3-C; uncharacterized LOC408365; chondroitin proteoglycan-2-like; cuticular protein analogous to peritrophins 3-A; uncharacterized LOC100576184; uncharacterized LOC727129; uncharacterized protein DDB_G0287625-like; uncharacterized LOC413560; uncharacterized LOC100577513 |
| GO: Molecular function | GO:0102336 | 0.0097652 | 0.0863622 | 1.605581 | 100578829 552205 725031 725255 | 3-oxo-arachidoyl-CoA synthase activity | elongation of very long chain fatty acids protein 1-like; elongation of very long chain fatty acids protein AAEL008004; elongation of very long chain fatty acids protein 6; elongation of very long chain fatty acids protein 7 |
| GO: Molecular function | GO:0102337 | 0.0097652 | 0.0863622 | 1.605581 | 100578829 552205 725031 725255 | 3-oxo-cerotoyl-CoA synthase activity | elongation of very long chain fatty acids protein 1-like; elongation of very long chain fatty acids protein AAEL008004; elongation of very long chain fatty acids protein 6; elongation of very long chain fatty acids protein 7 |
| GO: Molecular function | GO:0102338 | 0.0097652 | 0.0863622 | 1.605581 | 100578829 552205 725031 725255 | 3-oxo-lignoceronyl-CoA synthase activity | elongation of very long chain fatty acids protein 1-like; elongation of very long chain fatty acids protein AAEL008004; elongation of very long chain fatty acids protein 6; elongation of very long chain fatty acids protein 7 |
| GO: Molecular function | GO:0102756 | 0.0097652 | 0.0863622 | 1.605581 | 100578829 552205 725031 725255 | very-long-chain 3-ketoacyl-CoA synthase activity | elongation of very long chain fatty acids protein 1-like; elongation of very long chain fatty acids protein AAEL008004; elongation of very long chain fatty acids protein 6; elongation of very long chain fatty acids protein 7 |
| GO: Molecular function | GO:0016614 | 0.0231687 | 0.1211240 | 1.527243 | 552457 413098 552425 410745 | oxidoreductase activity, acting on CH-OH group of donors | glucose dehydrogenase [FAD, quinone]; glucose dehydrogenase [FAD, quinone]; glucose dehydrogenase [FAD, quinone]; glucose dehydrogenase [FAD, quinone] |
| GO: Molecular function | GO:0043565 | 0.0305000 | 0.1343292 | 1.305352 | 726162 724196 725495 726370 725220 725302 413391 724373 100576194 410643 724456 552099 724301 100576220 413060 413558 406077 724412 724238 408443 724796 724422 724297 725502 100576147 410657 | sequence-specific DNA binding | homeobox protein goosecoid; retinal homeobox protein Rx1; homeobox protein GBX-2; uncharacterized LOC726370; homeobox protein Nkx-6.1-like; H2.0-like homeobox protein; homeobox protein MSX-2; homeobox protein Nkx-2.5-like; forkhead box protein J1-B-like; homeobox protein abdominal-A homolog; homeobox protein Msx; homeotic protein empty spiracles; paired mesoderm homeobox protein 2-like; homeobox protein rough-like; DNA-binding protein D-ETS-4; photoreceptor-specific nuclear receptor; homeotic protein antennapedia; muscle segmentation homeobox; paired box protein Pax-6-like; uncharacterized LOC408443; homeobox protein SIX6; homeobox protein Hox-B1a; uncharacterized LOC724297; homeobox protein ARX-like; fork head domain-containing protein FD4; inhibitory POU protein |
| GO: Molecular function | GO:0004252 | 0.0451943 | 0.1393728 | 1.394571 | 726126 410438 724145 413645 724917 410894 724308 411358 409143 726718 | serine-type endopeptidase activity | proclotting enzyme; neuroendocrine convertase 1-like; transmembrane protease serine 9; trypsin alpha-3; uncharacterized LOC724917; chymotrypsin-1; trypsin-1; trypsin-1; venom serine protease 34; rhomboid-related protein 4-like |
| GO: Molecular function | GO:0004888 | 0.0468442 | 0.1396648 | 1.460218 | 410478 406148 100576745 726214 406153 | transmembrane signaling receptor activity | nicotinic acetylcholine receptor beta1 subunit; nicotinic acetylcholine receptor alpha7 subunit; leucine-rich repeat-containing protein 26-like; nicotinic acetylcholine receptor alpha1 subunit; nicotinic acetylcholine receptor alpha2 subunit |
| KEGG | KEGG:00900 | 0.0029130 | 0.0455160 | 1.666599 | 413763 551189 725363 725817 | Terpenoid backbone biosynthesis | hydroxymethylglutaryl-CoA synthase 1; farnesyl pyrophosphate synthase-like; isopentenyl-diphosphate Delta-isomerase 1; diphosphomevalonate decarboxylase |
| KEGG | KEGG:00910 | 0.0149136 | 0.0980780 | 1.563268 | 100577841 726898 727237 | Nitrogen metabolism | carbonic anhydrase 7-like; carbonic anhydrase 3; carbonic anhydrase 2-like |
| KEGG | KEGG:03022 | 0.0204082 | 0.1133787 | -1.375002 | 411786 412010 409906 550692 412268 410753 550665 551734 410305 410004 409735 411231 411144 411742 412919 410459 552563 551470 550895 724502 | Basal transcription factors | general transcription factor IIE subunit 1; general transcription factor IIH subunit 4; transcription initiation factor TFIID subunit 2; TATA-box-binding protein; transcription initiation factor IIA subunit 1; TATA-box-binding protein-like; transcription initiation factor IIA subunit 2; DNA excision repair protein haywire; transcription initiation factor TFIID subunit 12; transcription initiation factor TFIID subunit 10-like; transcription initiation factor TFIID subunit 6; general transcription factor IIH subunit 3; transcription initiation factor TFIID subunit 11; transcription initiation factor TFIID subunit 8-like; TATA box-binding protein-like protein 1; cyclin-H; transcription initiation factor TFIID subunit 7; general transcription factor IIF subunit 2; general transcription factor IIH subunit 1; transcription initiation factor TFIID subunit 5 |
| KEGG | KEGG:00360 | 0.0333296 | 0.1343292 | 1.499087 | 410639 725400 408622 410638 | Phenylalanine metabolism | alpha-methyldopa hypersensitive protein-like; 4-hydroxyphenylpyruvate dioxygenase; protein henna; aromatic-L-amino-acid decarboxylase |
Lasius flavus
Table S20: The results of GSEA (gene set enrichment analysis) for pheromone sensitivity in gene expression in Lasius flavus. The table lists GO and KEGG terms with their NES (normalized enrichment score), the associated raw and adjusted p-values (adjustment was performed using Benjamini-Hochberg correction), and the genes underlying the enrichment result.
tab_S20 <- GO_and_KEGG_results_GSEA %>%
filter(Species == "lf" & pvalue < 0.05) %>%
arrange(Test_type, pvalue) %>%
select(-Species, -ES, -nMoreExtreme, -size) %>%
add_name_col()
saveRDS(tab_S20 %>% select(-enriched_gene_ids, -gene_names), file = "supplement/tab_S20.rds")
kable.table(tab_S20)| Test_type | ID | pvalue | p.adjust | NES | enriched_gene_ids | Description | gene_names |
|---|---|---|---|---|---|---|---|
| GO: Biological process | GO:0006464 | 0.0079458 | 0.0827686 | 1.603380 | 551967 410872 | cellular protein modification process | probable tubulin polyglutamylase TTLL2; SUMO-activating enzyme subunit 1 |
| GO: Biological process | GO:0006364 | 0.0144071 | 0.0976497 | 1.597910 | 725143 411026 411233 | rRNA processing | probable U3 small nucleolar RNA-associated protein 11; WD repeat-containing protein 36; U3 small nucleolar ribonucleoprotein protein MPP10 |
| GO: Biological process | GO:0042742 | 0.0153002 | 0.0980780 | 1.552196 | 406142 406143 | defense response to bacterium | hymenoptaecin; defensin 1 |
| GO: Biological process | GO:0006633 | 0.0161826 | 0.0984543 | 1.582131 | 724552 100578829 725031 552205 412166 100577192 413789 | fatty acid biosynthetic process | elongation of very long chain fatty acids protein AAEL008004-like; elongation of very long chain fatty acids protein 1-like; elongation of very long chain fatty acids protein 6; elongation of very long chain fatty acids protein AAEL008004; acyl-CoA Delta(11) desaturase; very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 2; elongation of very long chain fatty acids protein AAEL008004-like |
| GO: Biological process | GO:0007020 | 0.0252395 | 0.1237232 | 1.512303 | 726073 411508 | microtubule nucleation | gamma-tubulin complex component 3; tubulin gamma-2 chain |
| GO: Cellular component | GO:0005730 | 0.0023484 | 0.0419353 | 1.785677 | 725143 413404 724432 724129 | nucleolus | probable U3 small nucleolar RNA-associated protein 11; ribosome biogenesis protein WDR12 homolog; ribosomal RNA processing protein 36 homolog; pescadillo homolog |
| GO: Cellular component | GO:0035267 | 0.0069751 | 0.0792628 | 1.630456 | 726816 408575 | NuA4 histone acetyltransferase complex | ruvB-like 2; DNA methyltransferase 1-associated protein 1 |
| GO: Cellular component | GO:0032040 | 0.0144522 | 0.0976497 | 1.577040 | 725143 411026 413649 | small-subunit processome | probable U3 small nucleolar RNA-associated protein 11; WD repeat-containing protein 36; rRNA-processing protein FCF1 homolog |
| GO: Cellular component | GO:0005886 | 0.0220967 | 0.1200905 | 1.505998 | 725384 412740 725205 551848 411611 552785 551782 552546 | plasma membrane | odorant receptor 2; ligand-gated chloride channel homolog 3; odorant receptor 1; protocadherin-like wing polarity protein stan; tachykinin-like peptides receptor 99D; ligand-gated ion channel pHCl; bestrophin-4-like; protocadherin Fat 4 |
| GO: Cellular component | GO:0005856 | 0.0244862 | 0.1237232 | 1.514313 | 406154 412827 | cytoskeleton | profilin; Bardet-Biedl syndrome 2 protein homolog |
| GO: Cellular component | GO:0031011 | 0.0356894 | 0.1343292 | 1.478919 | 726816 100576104 | Ino80 complex | ruvB-like 2; INO80 complex subunit B |
| GO: Molecular function | GO:0004984 | 0.0127138 | 0.0922074 | 1.440536 | 725384 725205 | olfactory receptor activity | odorant receptor 2; odorant receptor 1 |
| GO: Molecular function | GO:0003779 | 0.0162086 | 0.0984543 | 1.542631 | 406154 411744 | actin binding | profilin; spectrin beta chain |
| GO: Molecular function | GO:0005549 | 0.0176439 | 0.1025808 | 1.562299 | 725384 406109 406100 725205 406102 | odorant binding | odorant receptor 2; odorant binding protein 6; odorant binding protein 5; odorant receptor 1; odorant binding protein 1 |
| GO: Molecular function | GO:0042302 | 0.0290093 | 0.1318605 | 1.516549 | 100577562 100577189 409345 724556 727578 724624 | structural constituent of cuticle | probable LIM domain-containing serine/threonine-protein kinase DDB_G0286997; cuticular protein 6; cuticular protein 3; cuticular protein 17; uncharacterized LOC727578; endocuticle structural glycoprotein SgAbd-1 |
| GO: Molecular function | GO:0036459 | 0.0315236 | 0.1343292 | -1.577174 | 411920 412114 409389 552660 410109 411572 411981 411362 408619 | thiol-dependent ubiquitinyl hydrolase activity | ubiquitin carboxyl-terminal hydrolase 22; ubiquitin carboxyl-terminal hydrolase 35-like; ubiquitin specific protease-like; ubiquitin carboxyl-terminal hydrolase 46; ubiquitin carboxyl-terminal hydrolase 34; ubiquitin carboxyl-terminal hydrolase 14; probable ubiquitin carboxyl-terminal hydrolase FAF-X; ubiquitin carboxyl-terminal hydrolase 8-like; ubiquitin carboxyl-terminal hydrolase 3-like |
| GO: Molecular function | GO:0005319 | 0.0344271 | 0.1343292 | 1.495474 | 406088 410793 551250 411955 408696 726783 | lipid transporter activity | vitellogenin; uncharacterized LOC410793; microsomal triglyceride transfer protein large subunit; uncharacterized LOC411955; uncharacterized LOC408696; vitellogenin-like |
| GO: Molecular function | GO:0102336 | 0.0396585 | 0.1343292 | 1.478584 | 724552 100578829 725031 552205 413789 | 3-oxo-arachidoyl-CoA synthase activity | elongation of very long chain fatty acids protein AAEL008004-like; elongation of very long chain fatty acids protein 1-like; elongation of very long chain fatty acids protein 6; elongation of very long chain fatty acids protein AAEL008004; elongation of very long chain fatty acids protein AAEL008004-like |
| GO: Molecular function | GO:0102337 | 0.0396585 | 0.1343292 | 1.478584 | 724552 100578829 725031 552205 413789 | 3-oxo-cerotoyl-CoA synthase activity | elongation of very long chain fatty acids protein AAEL008004-like; elongation of very long chain fatty acids protein 1-like; elongation of very long chain fatty acids protein 6; elongation of very long chain fatty acids protein AAEL008004; elongation of very long chain fatty acids protein AAEL008004-like |
| GO: Molecular function | GO:0102338 | 0.0396585 | 0.1343292 | 1.478584 | 724552 100578829 725031 552205 413789 | 3-oxo-lignoceronyl-CoA synthase activity | elongation of very long chain fatty acids protein AAEL008004-like; elongation of very long chain fatty acids protein 1-like; elongation of very long chain fatty acids protein 6; elongation of very long chain fatty acids protein AAEL008004; elongation of very long chain fatty acids protein AAEL008004-like |
| GO: Molecular function | GO:0102756 | 0.0396585 | 0.1343292 | 1.478584 | 724552 100578829 725031 552205 413789 | very-long-chain 3-ketoacyl-CoA synthase activity | elongation of very long chain fatty acids protein AAEL008004-like; elongation of very long chain fatty acids protein 1-like; elongation of very long chain fatty acids protein 6; elongation of very long chain fatty acids protein AAEL008004; elongation of very long chain fatty acids protein AAEL008004-like |
| KEGG | KEGG:00130 | 0.0064743 | 0.0773036 | 1.632588 | 412082 725400 | Ubiquinone and other terpenoid-quinone biosynthesis | ubiquinone biosynthesis O-methyltransferase, mitochondrial; 4-hydroxyphenylpyruvate dioxygenase |
| KEGG | KEGG:04080 | 0.0118545 | 0.0922074 | 1.560081 | 410435 412740 413997 411323 412299 411611 412818 410654 406079 | Neuroactive ligand-receptor interaction | serotonin receptor; ligand-gated chloride channel homolog 3; translocator protein; serotonin receptor; muscarinic acetylcholine receptor DM1; tachykinin-like peptides receptor 99D; NMDA receptor 2; cholecystokinin receptor-like; NMDA receptor 1 |
| KEGG | KEGG:00061 | 0.0370928 | 0.1343292 | 1.479387 | 411959 409515 552286 551837 412815 | Fatty acid biosynthesis | fatty acid synthase-like; long-chain-fatty-acid–CoA ligase 4; acetyl-CoA carboxylase; long-chain-fatty-acid–CoA ligase ACSBG2; fatty acid synthase |
| KEGG | KEGG:00640 | 0.0382217 | 0.1343292 | 1.427875 | 409150 413942 551958 409624 410325 552286 551403 551208 | Propanoate metabolism | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial; acyl-CoA synthetase short-chain family member 3, mitochondrial; uncharacterized LOC551958; acetyl-coenzyme A synthetase; trifunctional enzyme subunit alpha, mitochondrial; acetyl-CoA carboxylase; succinyl-CoA ligase subunit alpha, mitochondrial; 3-hydroxyisobutyrate dehydrogenase, mitochondrial |
| KEGG | KEGG:00790 | 0.0383548 | 0.1343292 | 1.466181 | 412015 551072 | Folate biosynthesis | 6-pyruvoyl tetrahydrobiopterin synthase; carbonyl reductase [NADPH] 1-like |
| KEGG | KEGG:00630 | 0.0495742 | 0.1441111 | 1.413485 | 411541 409624 443552 411796 409485 | Glyoxylate and dicarboxylate metabolism | glycerate kinase; acetyl-coenzyme A synthetase; catalase; serine hydroxymethyltransferase; cytoplasmic aconitate hydratase-like |
Lasius niger
Table S21: The results of GSEA (gene set enrichment analysis) for pheromone sensitivity in gene expression in Lasius niger. The table lists GO and KEGG terms with their NES (normalized enrichment score), the associated raw and adjusted p-values (adjustment was performed using Benjamini-Hochberg correction), and the genes underlying the enrichment result.
tab_S21 <- GO_and_KEGG_results_GSEA %>%
filter(Species == "ln" & pvalue < 0.05) %>%
arrange(Test_type, pvalue) %>%
select(-Species, -ES, -nMoreExtreme, -size) %>%
add_name_col()
saveRDS(tab_S21 %>% select(-enriched_gene_ids, -gene_names), file = "supplement/tab_S21.rds")
kable.table(tab_S21)| Test_type | ID | pvalue | p.adjust | NES | enriched_gene_ids | Description | gene_names |
|---|---|---|---|---|---|---|---|
| GO: Biological process | GO:0030163 | 0.0020053 | 0.0419353 | -1.883620 | 551128 409198 410026 551343 551386 | protein catabolic process | 26S protease regulatory subunit 4; 26S protease regulatory subunit 6A-B; 26S proteasome regulatory complex subunit p48A; 26S protease regulatory subunit 7; 26S protease regulatory subunit 10B |
| GO: Biological process | GO:0009058 | 0.0397614 | 0.1343292 | 1.449365 | 411916 724239 411959 | biosynthetic process | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial; kynurenine/alpha-aminoadipate aminotransferase, mitochondrial-like; fatty acid synthase-like |
| GO: Cellular component | GO:0000139 | 0.0033846 | 0.0497738 | 1.691075 | 410030 100576099 408983 725057 | Golgi membrane | protein SEC13 homolog; carbohydrate sulfotransferase 11-like; protein transport protein Sec23A; coatomer subunit beta |
| GO: Cellular component | GO:0005576 | 0.0412082 | 0.1365156 | 1.321417 | 725217 726375 724246 410884 409241 410065 100577576 406083 409314 413481 413560 724880 551597 726057 724563 | extracellular region | uncharacterized protein PFB0145c-like; bone morphogenetic protein 2-B; growth/differentiation factor 2-like; phospholipase A1; prohormone-4; cuticular protein analogous to peritrophins 3-D; chondroitin proteoglycan-2-like; tachykinin; prohormone-2; probable chitinase 3; uncharacterized LOC413560; allatostatin A; cysteine-rich venom protein-like; insulin-like peptide 2; uncharacterized LOC724563 |
| GO: Cellular component | GO:0005680 | 0.0420468 | 0.1365156 | -1.494161 | 413499 413293 409810 | anaphase-promoting complex | cell division cycle protein 23 homolog; anaphase-promoting complex subunit 10; anaphase-promoting complex subunit 4 |
| GO: Molecular function | GO:0036402 | 0.0014025 | 0.0350631 | -1.880150 | 551128 409198 410026 551343 551386 | proteasome-activating ATPase activity | 26S protease regulatory subunit 4; 26S protease regulatory subunit 6A-B; 26S proteasome regulatory complex subunit p48A; 26S protease regulatory subunit 7; 26S protease regulatory subunit 10B |
| GO: Molecular function | GO:0008146 | 0.0060634 | 0.0773036 | 1.608466 | 412996 100576099 | sulfotransferase activity | estrogen sulfotransferase; carbohydrate sulfotransferase 11-like |
| GO: Molecular function | GO:0042302 | 0.0074711 | 0.0812075 | 1.587413 | 727197 726995 724624 724777 100576341 100577562 100577189 | structural constituent of cuticle | cuticular protein 2; endocuticle structural glycoprotein ABD-4; endocuticle structural glycoprotein SgAbd-1; cuticular protein 14; cuticular protein 16; probable LIM domain-containing serine/threonine-protein kinase DDB_G0286997; cuticular protein 6 |
| GO: Molecular function | GO:0005198 | 0.0100180 | 0.0863622 | 1.587925 | 410030 550716 725057 | structural molecule activity | protein SEC13 homolog; clathrin heavy chain; coatomer subunit beta |
| GO: Molecular function | GO:0005509 | 0.0251975 | 0.1237232 | 1.349267 | 408672 410368 100577629 411647 408405 413977 409061 408937 551313 411659 409762 551859 551661 409881 409367 412825 | calcium ion binding | programmed cell death protein 6; cadherin-23; uncharacterized LOC100577629; calumenin; uncharacterized LOC408405; calsyntenin-1; tyrosine kinase receptor Cad96Ca; SPARC; calcineurin subunit B type 2; troponin C type I; sarcoplasmic calcium-binding protein 1; calmodulin; uncharacterized LOC551661; myosin regulatory light chain 2; calcyphosin-like protein; sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 |
| GO: Molecular function | GO:0036459 | 0.0307918 | 0.1343292 | -1.532409 | 411981 411572 410109 411362 409389 552660 408619 412114 | thiol-dependent ubiquitinyl hydrolase activity | probable ubiquitin carboxyl-terminal hydrolase FAF-X; ubiquitin carboxyl-terminal hydrolase 14; ubiquitin carboxyl-terminal hydrolase 34; ubiquitin carboxyl-terminal hydrolase 8-like; ubiquitin specific protease-like; ubiquitin carboxyl-terminal hydrolase 46; ubiquitin carboxyl-terminal hydrolase 3-like; ubiquitin carboxyl-terminal hydrolase 35-like |
| GO: Molecular function | GO:0008536 | 0.0457143 | 0.1393728 | -1.483124 | 413940 413344 413636 412817 726133 409992 411865 | Ran GTPase binding | importin-9; exportin-5; exportin-7; importin-4-like; exportin-6; importin-13; exportin-2 |
| KEGG | KEGG:01040 | 0.0020385 | 0.0419353 | 1.691761 | 412166 725031 552417 725146 | Biosynthesis of unsaturated fatty acids | acyl-CoA Delta(11) desaturase; elongation of very long chain fatty acids protein 6; acyl-CoA Delta(11) desaturase; very-long-chain enoyl-CoA reductase |
| KEGG | KEGG:03050 | 0.0129090 | 0.0922074 | 1.475489 | 551550 409880 409609 409168 411695 409699 409668 | Proteasome | probable 26S proteasome non-ATPase regulatory subunit 3; 26S proteasome non-ATPase regulatory subunit 12; 26S proteasome non-ATPase regulatory subunit 4; 26S proteasome non-ATPase regulatory subunit 13; proteasome subunit beta type-1; 26S proteasome non-ATPase regulatory subunit 1-like; proteasome inhibitor PI31 subunit |
| KEGG | KEGG:04080 | 0.0268860 | 0.1254808 | 1.423729 | 408995 412740 551388 411760 412011 411420 406079 726970 552518 412299 | Neuroactive ligand-receptor interaction | D2-like dopamine receptor; ligand-gated chloride channel homolog 3; adipokinetic hormone receptor; metabotropic glutamate receptor 7; probable muscarinic acetylcholine receptor gar-2; adenosine receptor A2b; NMDA receptor 1; cys-loop ligand-gated ion channel subunit 8916; serotonin receptor; muscarinic acetylcholine receptor DM1 |
| KEGG | KEGG:00260 | 0.0469274 | 0.1396648 | 1.401367 | 412674 411916 406081 552832 411796 411541 | Glycine, serine and threonine metabolism | phosphoserine phosphatase; 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial; glucose oxidase; glycine N-methyltransferase; serine hydroxymethyltransferase; glycerate kinase |
GO and KEGG enrichment for differentially spliced genes
This section uses a metric that measures the sensitivity of alternative splicing to pheromone treatment (referred to in the code below as sensitivity). This metric is calculated by finding the highest and lowest log fold change in expression among the isoforms for gene \(i\), and then taking the difference. For example, a gene with two isoform, with log fold change values of -1 and +3, would have a sensitivity value of 4. In the following code, we calculate sensitivity for all of the genes which A) have orthologs in all 4 species, and B) have 2 or more isoforms. We then calculate the Spearman correlation in sensitivity across genes for each species pair, and also perform GSEA (as was previously done for the data on pheromone sensitivity).
# Calculate splicing sensitivity for Apis:
am_isoforms <- tbl(my_db, "ebseq_isoform_am") %>%
left_join(tbl(my_db, "isoforms_am"), by = "isoform") %>%
arrange(gene) %>% collect()
# Discard genes with only one splice variant
keep <- am_isoforms %>% group_by(gene) %>% summarise(n_isoforms = n()) %>% filter(n_isoforms > 1) %>% .$gene
am_isoforms <- am_isoforms %>% filter(gene %in% keep) %>%
select(gene, PPDE, PostFC)
# Splicing index is max - min of the log(FC) for the isoforms of the focal gene
am.splice <- am_isoforms %>%
split(.$gene) %>%
purrr::map_dbl(function(x){
max(log(x$PostFC)) - min(log(x$PostFC))
})
am.splice <- data.frame(gene = names(am.splice),
sensitivity = unname(am.splice), stringsAsFactors = FALSE) %>%
arrange(-sensitivity)
# Calculate splicing sensitivity for the other 3 species. Here, there is an extra step: map the genes to their Apis orthologs
get_splice_score <- function(sp){
tabl <- paste("ebseq_isoform_", sp, sep = "")
iso.table <- paste("isoforms_", sp, sep = "")
ortho.table <- make.OGGs(c("am", sp))[[2]]
isoforms <- tbl(my_db, tabl) %>%
left_join(tbl(my_db, iso.table), by = "isoform") %>% collect(n=Inf) %>%
arrange(gene) %>%
left_join(ortho.table, by = c("gene" = sp)) %>%
select(-gene) %>% dplyr::rename(gene = am) %>%
filter(!is.na(gene)) %>% collect()
keep <- isoforms %>% group_by(gene) %>% summarise(n_isoforms = n()) %>% filter(n_isoforms > 1) %>% .$gene
isoforms <- isoforms %>% filter(gene %in% keep) %>%
select(gene, PPDE, PostFC)
isoforms <- isoforms %>%
split(.$gene) %>%
purrr::map_dbl(function(x){
max(log(x$PostFC)) - min(log(x$PostFC))
})
data.frame(gene = names(isoforms),
sensitivity = unname(isoforms), stringsAsFactors = FALSE) %>%
arrange(-sensitivity)
}
# Get the splicing index for all 4 species, and restrict to the set of genes that have orthologs in all 4 species
splice_scores <- list(am.splice,
get_splice_score("bt"),
get_splice_score("lf"),
get_splice_score("ln"))
# Find the correlations between each species pair in the splicing index
splicing_correlations <- data.frame(t(combn(1:4, 2)), rho = 0, p = 0) %>%
dplyr::rename(Species1 = X1, Species2 = X2)
for(i in 1:nrow(splicing_correlations)){
focal <- left_join(splice_scores[[splicing_correlations[i, 1]]],
splice_scores[[splicing_correlations[i, 2]]], by="gene")
focal <- focal[complete.cases(focal), ]
test <- with(focal, cor.test(sensitivity.x, sensitivity.y, method = "spearman"))
splicing_correlations$rho[i] <- test$estimate
splicing_correlations$p[i] <- test$p.value
}
splicing_correlations[,1] <- c("Apis mellifera", "Bombus terrestris", "Lasius flavus", "Lasius niger")[splicing_correlations[,1]]
splicing_correlations[,2] <- c("Apis mellifera", "Bombus terrestris", "Lasius flavus", "Lasius niger")[splicing_correlations[,2]]
splicing_correlations$p.adjust <- p.adjust(splicing_correlations$p, method = "BH")
splicing_correlations$sig <- " "
splicing_correlations$sig[splicing_correlations$p.adjust < 0.05] <- "*"
# Perform GSEA on the splicing index
GO_and_KEGG_splicing_GSEA <- rbind(
GO.and.KEGG.gsea(df = splice_scores[[1]], keep.all = TRUE) %>% mutate(Species = "am"),
GO.and.KEGG.gsea(df = splice_scores[[2]], keep.all = TRUE) %>% mutate(Species = "bt"),
GO.and.KEGG.gsea(df = splice_scores[[3]], keep.all = TRUE) %>% mutate(Species = "lf"),
GO.and.KEGG.gsea(df = splice_scores[[4]], keep.all = TRUE) %>% mutate(Species = "ln")) %>%
fill_in_GSEA_results() Inter-species correlations in the sensitivity of alternative splicing to queen pheromone across pairs of orthologous genes
Table S22: The table shows the Spearman correlation (rho) and p-value for correlations across genes in the pheromone-sensitivity of their isoform production, for each pair of species. For each gene, our metric of the sensitivity of splicing to pheromone treatment was calculated by taking the difference between the highest and lowest log fold change values for the various isoforms. Thus, genes for which one isoform strongly increased in expression and one strongly decreased following pheromone treatment score high, and those in which there is no response to pheromone – or a consistent response for all isoforms – score low. The results suggest that the pheromone sensitivity in splicing is highly conserved between orthologous bee genes, and somewhat less conserved between orthologous ant genes, and between bee and ants genes.
saveRDS(splicing_correlations, file = "supplement/tab_S22.rds")
splicing_correlations %>% pander(split.cell = 40, split.table = Inf)| Species1 | Species2 | rho | p | p.adjust | sig |
|---|---|---|---|---|---|
| Apis mellifera | Bombus terrestris | 0.1855 | 1.155e-08 | 6.927e-08 | * |
| Apis mellifera | Lasius flavus | 0.06259 | 0.06836 | 0.1367 | |
| Apis mellifera | Lasius niger | 0.04333 | 0.153 | 0.1836 | |
| Bombus terrestris | Lasius flavus | 0.08915 | 0.01015 | 0.03045 | * |
| Bombus terrestris | Lasius niger | 0.03039 | 0.2936 | 0.2936 | |
| Lasius flavus | Lasius niger | 0.04089 | 0.1499 | 0.1836 |
GSEA for pheromone sensitivity of alternative splicing
fig_S5 <- GO_and_KEGG_splicing_GSEA %>% make_enrichment_heatmap()
saveRDS(fig_S5, file = "supplement/fig_S5.rds")
fig_S5 %>% grid.draw()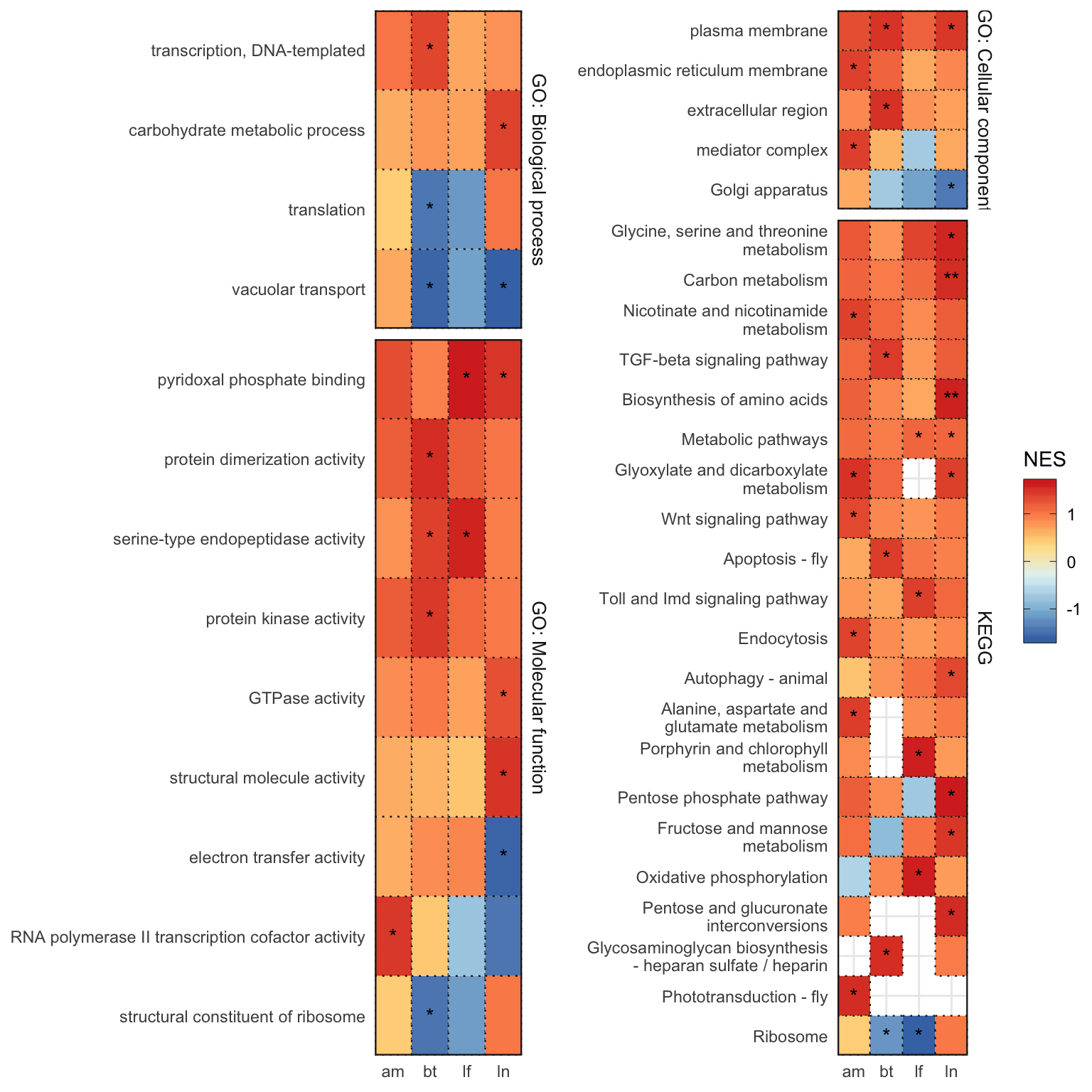
Figure S5: Genes for which alternative splicing is strongly affected by queen pheromone tend to have similar Gene Ontology and KEGG terms in ants and bees, although the data do not provide strong evidence for or against inter-species similarity. The colour shows the normalised expression score from a GSEA (gene set enrichment analysis) test implemented in the R package fgsea; positive (red) values indicate that the GO or KEGG term is over-represented among genes whose splicing is strongly affected by queen pheromone, and negative (blue) values indicate under-representation among those genes. Asterisks denote statistically significant enrichment (p < 0.05), and double asterisks mark results that remained significant after adjusting the p-values for multiple testing using the Benjamini-Hochberg method. Empty squares denote cases where we did not find at least 5 alternatively spliced genes annotated with the focal term.
Tabular results for GSEA for pheromone sensitivity of alternative splicing
Table S23: The results of GSEA (gene set enrichment analysis) for pheromone sensitivity in alternative splicing. The table lists statistically significant GO and KEGG terms with their NES (normalized enrichment score), the associated raw and adjusted p-values (adjustment was performed using Benjamini-Hochberg correction), and the genes underlying each enrichment result.
tab_S23 <- GO_and_KEGG_splicing_GSEA %>%
filter(pvalue < 0.05) %>%
arrange(Species, Test_type, pvalue) %>%
select(-ES, -nMoreExtreme, -size) %>%
add_name_col()
saveRDS(tab_S23 %>% select(-gene_names, -enriched_gene_ids), file = "supplement/tab_S23.rds")
kable.table(tab_S23)| Test_type | ID | pvalue | p.adjust | NES | enriched_gene_ids | Description | Species | gene_names |
|---|---|---|---|---|---|---|---|---|
| GO: Cellular component | GO:0005789 | 0.0371618 | 0.1390547 | 1.449603 | 411459 552377 413169 550724 | endoplasmic reticulum membrane | am | sterol regulatory element-binding protein cleavage-activating protein; diacylglycerol O-acyltransferase 1; presenilin-1; 3-hydroxy-3-methylglutaryl-coenzyme A reductase |
| GO: Cellular component | GO:0016592 | 0.0382692 | 0.1390547 | 1.452031 | 409749 550881 552719 | mediator complex | am | mediator of RNA polymerase II transcription subunit 31; mediator of RNA polymerase II transcription subunit 8; mediator of RNA polymerase II transcription subunit 11 |
| GO: Molecular function | GO:0001104 | 0.0322844 | 0.1273924 | 1.455521 | 409749 550881 552719 | RNA polymerase II transcription cofactor activity | am | mediator of RNA polymerase II transcription subunit 31; mediator of RNA polymerase II transcription subunit 8; mediator of RNA polymerase II transcription subunit 11 |
| KEGG | KEGG:04144 | 0.0044000 | 0.0725780 | 1.432046 | 409230 724991 413680 411585 411362 410923 413457 413464 411147 552297 551408 412601 409910 411723 409006 | Endocytosis | am | arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2; phosphatidylinositol 4-phosphate 5-kinase type-1 alpha; epsin-2; E3 ubiquitin-protein ligase NRDP1; ubiquitin carboxyl-terminal hydrolase 8-like; dynamin; zinc finger FYVE domain-containing protein 9; tyrosine-protein kinase Src64B; AP-2 complex subunit alpha; low density lipoprotein receptor adapter protein 1-B-like; uncharacterized LOC551408; mothers against decapentaplegic homolog 3; ras-like GTP-binding protein Rho1; E3 ubiquitin-protein ligase Nedd-4; arrestin red cell |
| KEGG | KEGG:04745 | 0.0071499 | 0.0869908 | 1.597421 | 409020 551691 550818 410489 | Phototransduction - fly | am | uncharacterized LOC409020; calcium/calmodulin-dependent protein kinase II; guanine nucleotide-binding protein G(q) subunit alpha; myosin-IIIb |
| KEGG | KEGG:00630 | 0.0121013 | 0.0981553 | 1.552847 | 413678 726754 | Glyoxylate and dicarboxylate metabolism | am | serine–pyruvate aminotransferase, mitochondrial; kynurenine formamidase |
| KEGG | KEGG:04310 | 0.0231533 | 0.1145620 | 1.393171 | 551691 410190 412108 409286 409158 411046 413169 409910 551517 552805 | Wnt signaling pathway | am | calcium/calmodulin-dependent protein kinase II; tyrosine-protein kinase Dnt; casein kinase I-like; stress-activated protein kinase JNK; C-terminal-binding protein; uncharacterized LOC411046; presenilin-1; ras-like GTP-binding protein Rho1; beta-TrCP; protein shifted |
| KEGG | KEGG:00250 | 0.0251192 | 0.1145620 | 1.478191 | 413372 413678 551113 408991 724480 | Alanine, aspartate and glutamate metabolism | am | putative glutamate synthase [NADPH]; serine–pyruvate aminotransferase, mitochondrial; delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial; glutaminase kidney isoform, mitochondrial; asparagine synthetase [glutamine-hydrolyzing] |
| KEGG | KEGG:00760 | 0.0390496 | 0.1390547 | 1.447503 | 551770 409487 408470 | Nicotinate and nicotinamide metabolism | am | cytosolic purine 5’-nucleotidase; probable glutamine-dependent NAD(+) synthetase; NAD kinase-like |
| GO: Biological process | GO:0007034 | 0.0205327 | 0.1145620 | -1.584054 | 550738 411857 410607 410883 | vacuolar transport | bt | NEDD4 family-interacting protein 1-like; charged multivesicular body protein 4b; charged multivesicular body protein 3; charged multivesicular body protein 6 |
| GO: Biological process | GO:0006351 | 0.0449834 | 0.1511387 | 1.363095 | 412010 410757 726204 409321 409227 411279 | transcription, DNA-templated | bt | general transcription factor IIH subunit 4; circadian locomoter output cycles protein kaput; hairy/enhancer-of-split related with YRPW motif protein 1; mothers against decapentaplegic homolog 4; ultraspiracle; actin-related protein 8 |
| GO: Biological process | GO:0006412 | 0.0476190 | 0.1511387 | -1.428849 | 724125 725943 552774 724631 413875 411862 550651 725147 413884 552097 552564 725854 552517 724708 412984 413137 409479 552106 412266 552676 | translation | bt | 28S ribosomal protein S7, mitochondrial; ubiquitin-60S ribosomal protein L40; 60S ribosomal protein L28; 60S ribosomal protein L29; 60S ribosomal protein L31; 28S ribosomal protein S2, mitochondrial; 40S ribosomal protein S4; 40S ribosomal protein S29; ubiquitin-40S ribosomal protein S27a; 39S ribosomal protein L14, mitochondrial; 40S ribosomal protein S7; 39S ribosomal protein L37, mitochondrial; 60S ribosomal protein L13; 39S ribosomal protein L23, mitochondrial; 28S ribosomal protein S30, mitochondrial; 60S ribosomal protein L15; 60S ribosomal protein L6; uncharacterized LOC552106; 60S ribosomal protein L27; 39S ribosomal protein L3, mitochondrial |
| GO: Cellular component | GO:0005886 | 0.0032006 | 0.0725780 | 1.537862 | 100578210 100577938 100577446 100577755 725297 100578083 100577334 411760 100578230 725205 551782 | plasma membrane | bt | odorant receptor 5; odorant receptor 18; odorant receptor 27; odorant receptor 30a-like; gustatory receptor 10; odorant receptor 14; odorant receptor 33; metabotropic glutamate receptor 7; odorant receptor 115; odorant receptor 1; bestrophin-4-like |
| GO: Cellular component | GO:0005576 | 0.0110065 | 0.0977523 | 1.562965 | 409307 409553 | extracellular region | bt | uncharacterized LOC409307; pancreatic triacylglycerol lipase-like |
| GO: Molecular function | GO:0046983 | 0.0110406 | 0.0977523 | 1.535076 | 410757 726204 410953 | protein dimerization activity | bt | circadian locomoter output cycles protein kaput; hairy/enhancer-of-split related with YRPW motif protein 1; MLX-interacting protein |
| GO: Molecular function | GO:0003735 | 0.0227273 | 0.1145620 | -1.444564 | 724125 725943 552774 724631 413875 411862 550651 725147 413884 552097 552564 725854 552517 724708 412984 413137 409479 552106 412266 552676 | structural constituent of ribosome | bt | 28S ribosomal protein S7, mitochondrial; ubiquitin-60S ribosomal protein L40; 60S ribosomal protein L28; 60S ribosomal protein L29; 60S ribosomal protein L31; 28S ribosomal protein S2, mitochondrial; 40S ribosomal protein S4; 40S ribosomal protein S29; ubiquitin-40S ribosomal protein S27a; 39S ribosomal protein L14, mitochondrial; 40S ribosomal protein S7; 39S ribosomal protein L37, mitochondrial; 60S ribosomal protein L13; 39S ribosomal protein L23, mitochondrial; 28S ribosomal protein S30, mitochondrial; 60S ribosomal protein L15; 60S ribosomal protein L6; uncharacterized LOC552106; 60S ribosomal protein L27; 39S ribosomal protein L3, mitochondrial |
| GO: Molecular function | GO:0004672 | 0.0267296 | 0.1145620 | 1.453553 | 408664 408533 413759 413190 551773 412747 | protein kinase activity | bt | homeodomain-interacting protein kinase 2; mitogen-activated protein kinase kinase kinase 15; SCY1-like protein 2; calcium-dependent protein kinase 4-like; serine/threonine-protein kinase VRK1-like; receptor interacting protein kinase 5 |
| GO: Molecular function | GO:0004252 | 0.0466352 | 0.1511387 | 1.396669 | 726126 410438 724145 413645 724917 410894 724308 411358 409143 726718 | serine-type endopeptidase activity | bt | proclotting enzyme; neuroendocrine convertase 1-like; transmembrane protease serine 9; trypsin alpha-3; uncharacterized LOC724917; chymotrypsin-1; trypsin-1; trypsin-1; venom serine protease 34; rhomboid-related protein 4-like |
| KEGG | KEGG:00534 | 0.0049711 | 0.0725780 | 1.595536 | 551445 413271 408293 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | bt | heparin sulfate O-sulfotransferase; galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase I; exostosin-1 |
| KEGG | KEGG:04214 | 0.0177298 | 0.1145620 | 1.471707 | 725245 551969 409227 409286 408533 408758 | Apoptosis - fly | bt | protein eiger-like; serine protease HTRA2, mitochondrial; ultraspiracle; stress-activated protein kinase JNK; mitogen-activated protein kinase kinase kinase 15; ecdysteroid-regulated gene E74 |
| KEGG | KEGG:04350 | 0.0207798 | 0.1145620 | 1.488501 | 410035 411627 409321 412866 | TGF-beta signaling pathway | bt | dorsal-ventral patterning protein Sog; retinoblastoma-like protein 1; mothers against decapentaplegic homolog 4; E3 ubiquitin-protein ligase SMURF2 |
| KEGG | KEGG:03010 | 0.0370370 | 0.1390547 | -1.340568 | 724125 725943 552774 724631 413875 409552 411862 550651 725147 552097 725168 552564 552517 724708 413137 409479 552106 412266 552676 | Ribosome | bt | 28S ribosomal protein S7, mitochondrial; ubiquitin-60S ribosomal protein L40; 60S ribosomal protein L28; 60S ribosomal protein L29; 60S ribosomal protein L31; 40S ribosomal protein S10-like; 28S ribosomal protein S2, mitochondrial; 40S ribosomal protein S4; 40S ribosomal protein S29; 39S ribosomal protein L14, mitochondrial; ribosomal protein S14; 40S ribosomal protein S7; 60S ribosomal protein L13; 39S ribosomal protein L23, mitochondrial; 60S ribosomal protein L15; 60S ribosomal protein L6; uncharacterized LOC552106; 60S ribosomal protein L27; 39S ribosomal protein L3, mitochondrial |
| GO: Molecular function | GO:0030170 | 0.0048622 | 0.0725780 | 1.641783 | 724919 411796 408509 408817 410583 408432 | pyridoxal phosphate binding | lf | mitochondrial amidoxime reducing component 2-like; serine hydroxymethyltransferase; glutamate decarboxylase 1; alanine–glyoxylate aminotransferase 2-like; ornithine aminotransferase, mitochondrial; glutamate decarboxylase |
| GO: Molecular function | GO:0004252 | 0.0079400 | 0.0891724 | 1.592918 | 725154 409459 412319 409204 412293 | serine-type endopeptidase activity | lf | serine protease snake; lon protease homolog, mitochondrial; rhomboid-related protein 2; trypsin; membrane-bound transcription factor site-1 protease |
| KEGG | KEGG:00190 | 0.0024240 | 0.0725780 | 1.681785 | 409103 408367 | Oxidative phosphorylation | lf | protoheme IX farnesyltransferase, mitochondrial; NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial |
| KEGG | KEGG:00860 | 0.0026977 | 0.0725780 | 1.681653 | 409103 494506 409922 | Porphyrin and chlorophyll metabolism | lf | protoheme IX farnesyltransferase, mitochondrial; heme oxygenase; ferrochelatase, mitochondrial |
| KEGG | KEGG:03010 | 0.0113821 | 0.0977523 | -1.827766 | 724125 411103 411380 413398 409832 552517 413868 725201 725062 | Ribosome | lf | 28S ribosomal protein S7, mitochondrial; putative 28S ribosomal protein S5, mitochondrial; 60S ribosomal protein L30; 39S ribosomal protein L32, mitochondrial; 60S ribosomal protein L18a; 60S ribosomal protein L13; 60S ribosomal protein L10a; 39S ribosomal protein L21, mitochondrial; 39S ribosomal protein L4, mitochondrial |
| KEGG | KEGG:04624 | 0.0274635 | 0.1145620 | 1.459957 | 406086 725832 551608 725154 724703 | Toll and Imd signaling pathway | lf | dorsal; beta-1,3-glucan-binding protein 1; serine/threonine-protein kinase pelle; serine protease snake; coagulation factor X |
| KEGG | KEGG:01100 | 0.0467953 | 0.1511387 | 1.162914 | 409444 409103 413119 409276 494506 550970 408441 412731 408299 552086 411140 410828 727004 100577378 552823 551853 409515 727189 411796 552771 409499 100577053 726824 412170 408509 409614 409329 410330 408817 552522 413255 551419 413411 412541 412548 411633 411662 410583 551208 408432 411372 409224 410080 552007 552421 727456 413664 551749 410530 411563 408730 552533 413643 551005 551041 408809 410554 725665 551403 410627 409250 552556 726216 409494 409270 408883 551578 550785 409922 724718 724550 412341 412815 412674 552130 406107 727300 410118 409861 410071 550885 551762 552657 410132 724361 724991 725817 413705 100577717 | Metabolic pathways | lf | AMP deaminase 2; protoheme IX farnesyltransferase, mitochondrial; heparan-alpha-glucosaminide N-acetyltransferase-like; phosphatidylinositol 5-phosphate 4-kinase type-2 alpha; heme oxygenase; lysophosphatidylcholine acyltransferase-like; adenosine kinase 1; type II inositol 1,4,5-trisphosphate 5-phosphatase; purine nucleoside phosphorylase; aldose 1-epimerase; putative aldehyde dehydrogenase family 7 member A1 homolog; tryptophan 2,3-dioxygenase; putative glutathione-specific gamma-glutamylcyclotransferase 2; galactokinase-like; malonyl-CoA decarboxylase, mitochondrial-like; STT3, subunit of the oligosaccharyltransferase complex, homolog B; long-chain-fatty-acid–CoA ligase 4; geranylgeranyl pyrophosphate synthase; serine hydroxymethyltransferase; hydroxyacid oxidase 1; UDP-glucose 4-epimerase-like; glycine dehydrogenase (decarboxylating), mitochondrial; putative inositol monophosphatase 3; putative neutral sphingomyelinase; glutamate decarboxylase 1; group XIIA secretory phospholipase A2; mannose-1-phosphate guanyltransferase beta; phosphatidylserine synthase 1; alanine–glyoxylate aminotransferase 2-like; alpha-(1,6)-fucosyltransferase; 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial; alkaline ceramidase; myotubularin-related protein 14; long-chain-fatty-acid–CoA ligase 6; 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial; UDP-glucose 4-epimerase; probable trans-2-enoyl-CoA reductase, mitochondrial; ornithine aminotransferase, mitochondrial; 3-hydroxyisobutyrate dehydrogenase, mitochondrial; glutamate decarboxylase; glutathione synthetase-like; ubiquinone biosynthesis protein COQ7; spermine synthase; pyruvate kinase; glycogenin-1; glucose-6-phosphate 1-epimerase; GPI transamidase component PIG-S; glycosyltransferase-like protein LARGE1; alkaline phosphatase-like; myotubularin-related protein 2; inositol polyphosphate 5-phosphatase K-like; GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase; N-acetylglucosaminyl-phosphatidylinositol biosynthetic protein; hexokinase type 2; glycerol kinase; diacylglycerol kinase theta; methylcrotonoyl-CoA carboxylase beta chain, mitochondrial; 5-oxoprolinase; succinyl-CoA ligase subunit alpha, mitochondrial; putative aminopeptidase W07G4.4; beta-ureidopropionase; arginase-1; exostosin-2; methylglutaconyl-CoA hydratase, mitochondrial; glycerol-3-phosphate acyltransferase 4; deoxycytidylate deaminase; protein O-mannosyl-transferase 2; fructose-bisphosphate aldolase; ferrochelatase, mitochondrial; xanthine dehydrogenase; iduronate 2-sulfatase; alpha-methylacyl-CoA racemase; fatty acid synthase; phosphoserine phosphatase; alpha-aminoadipic semialdehyde synthase, mitochondrial; glucosamine-fructose-6-phosphate aminotransferase 2; low molecular weight phosphotyrosine protein phosphatase-like; D-glucuronyl C5-epimerase; nucleoside diphosphate kinase; glycerol-3-phosphate phosphatase-like; nicotinate phosphoribosyltransferase; adenosylhomocysteinase 2-like; phosphoribosylformylglycinamidine synthase; GPI transamidase component PIG-T; GPI inositol-deacylase; phosphatidylinositol 4-phosphate 5-kinase type-1 alpha; diphosphomevalonate decarboxylase; acidic mammalian chitinase-like; uncharacterized LOC100577717 |
| GO: Biological process | GO:0007034 | 0.0242511 | 0.1145620 | -1.627514 | 410607 552786 550738 411857 410883 | vacuolar transport | ln | charged multivesicular body protein 3; charged multivesicular body protein 2a; NEDD4 family-interacting protein 1-like; charged multivesicular body protein 4b; charged multivesicular body protein 6 |
| GO: Biological process | GO:0005975 | 0.0289666 | 0.1174757 | 1.386429 | 551785 409267 411100 409884 552381 726210 551447 412362 726095 406114 411897 409199 412245 727456 | carbohydrate metabolic process | ln | 6-phosphogluconolactonase; glycogen phosphorylase; FGGY carbohydrate kinase domain-containing protein; xylulose kinase; neutral and basic amino acid transport protein rBAT; uncharacterized family 31 glucosidase KIAA1161; mannose-6-phosphate isomerase; glucose 1,6-bisphosphate synthase; galactoside 2-alpha-L-fucosyltransferase 2-like; alpha-amylase; phosphoglucomutase; glycerol kinase; acidic mammalian chitinase; glucose-6-phosphate 1-epimerase |
| GO: Cellular component | GO:0005794 | 0.0228188 | 0.1145620 | -1.587668 | 412710 411970 412913 409613 411408 | Golgi apparatus | ln | trafficking protein particle complex subunit 3; G kinase-anchoring protein 1-like; Golgi SNAP receptor complex member 2; GTP-binding protein SAR1; CDP-diacylglycerol–inositol 3-phosphatidyltransferase |
| GO: Cellular component | GO:0005886 | 0.0241376 | 0.1145620 | 1.511492 | 409650 551165 552552 406124 551508 725384 551388 | plasma membrane | ln | solute carrier organic anion transporter family member 2A1; innexin inx3; uncharacterized LOC552552; gamma-aminobutyric acid receptor subunit beta; vang-like protein 1; odorant receptor 2; adipokinetic hormone receptor |
| GO: Molecular function | GO:0030170 | 0.0185166 | 0.1145620 | 1.478569 | 411796 409267 409927 724919 | pyridoxal phosphate binding | ln | serine hydroxymethyltransferase; glycogen phosphorylase; putative pyridoxal-dependent decarboxylase domain-containing protein 2; mitochondrial amidoxime reducing component 2-like |
| GO: Molecular function | GO:0009055 | 0.0191898 | 0.1145620 | -1.590390 | 413605 727309 552386 409549 552835 410308 408270 551039 551169 551710 | electron transfer activity | ln | cytochrome c1, heme protein, mitochondrial; glutaredoxin-C4; anamorsin homolog; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; glutaredoxin-related protein 5, mitochondrial; electron transfer flavoprotein subunit beta; mitochondrial cytochrome C; dihydrolipoyl dehydrogenase, mitochondrial; succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial-like; electron transfer flavoprotein subunit alpha, mitochondrial |
| GO: Molecular function | GO:0005198 | 0.0268312 | 0.1145620 | 1.488230 | 550716 410030 725057 412083 | structural molecule activity | ln | clathrin heavy chain; protein SEC13 homolog; coatomer subunit beta; coatomer subunit gamma |
| GO: Molecular function | GO:0003924 | 0.0472047 | 0.1511387 | 1.289588 | 406098 411351 551731 409529 413943 550723 410241 410969 410906 551185 413034 411542 410414 413614 411472 409126 410157 | GTPase activity | ln | translation initiation factor 2; elongation factor G, mitochondrial; rho GTPase-activating protein 190; ras-like protein 2; eukaryotic peptide chain release factor GTP-binding subunit ERF3A; ras-related protein Rab-39B; ras-related protein Rab-10; ras-related protein Rab-9A; guanine nucleotide-binding protein subunit alpha homolog; GTP-binding protein 1; rho-related BTB domain-containing protein 1; signal recognition particle receptor subunit alpha homolog; 116 kDa U5 small nuclear ribonucleoprotein component; ras-related protein Rab-14; dynamin-1-like protein; ras-related protein Rab-2; ras-related protein Rab-23 |
| KEGG | KEGG:01230 | 0.0002005 | 0.0219000 | 1.671990 | 411796 413867 552007 412876 550785 550804 550767 408859 | Biosynthesis of amino acids | ln | serine hydroxymethyltransferase; transaldolase; pyruvate kinase; pyruvate carboxylase, mitochondrial; fructose-bisphosphate aldolase; transketolase; ribose 5-phosphate isomerase A; pyrroline-5-carboxylate reductase 2 |
| KEGG | KEGG:01200 | 0.0003000 | 0.0219000 | 1.597283 | 411796 551785 413867 552007 412876 409624 550785 550804 550767 550667 | Carbon metabolism | ln | serine hydroxymethyltransferase; 6-phosphogluconolactonase; transaldolase; pyruvate kinase; pyruvate carboxylase, mitochondrial; acetyl-coenzyme A synthetase; fructose-bisphosphate aldolase; transketolase; ribose 5-phosphate isomerase A; succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial |
| KEGG | KEGG:00030 | 0.0012452 | 0.0605998 | 1.711683 | 551785 413867 550785 550804 550767 | Pentose phosphate pathway | ln | 6-phosphogluconolactonase; transaldolase; fructose-bisphosphate aldolase; transketolase; ribose 5-phosphate isomerase A |
| KEGG | KEGG:00260 | 0.0035281 | 0.0725780 | 1.638785 | 411796 406081 410432 | Glycine, serine and threonine metabolism | ln | serine hydroxymethyltransferase; glucose oxidase; L-threonine 3-dehydrogenase, mitochondrial |
| KEGG | KEGG:00040 | 0.0061873 | 0.0821217 | 1.612270 | 551968 409884 | Pentose and glucuronate interconversions | ln | aldose reductase-like; xylulose kinase |
| KEGG | KEGG:04140 | 0.0108011 | 0.0977523 | 1.393260 | 552756 413663 409529 413153 408501 409393 726637 552038 412687 100578537 409941 413668 413430 551198 552315 551668 551057 724873 409577 | Autophagy - animal | ln | cathepsin L1; myotubularin-related protein 4; ras-like protein 2; uncharacterized LOC413153; serine/threonine-protein kinase/endoribonuclease IRE1; serine/threonine-protein kinase mTOR; ubiquitin-like modifier-activating enzyme atg7; synaptosomal-associated protein 29; beclin 1-associated autophagy-related key regulator; UV radiation resistance-associated gene protein; zinc finger FYVE domain-containing protein 1-like; run domain Beclin-1-interacting and cysteine-rich domain-containing protein; RAC serine/threonine-protein kinase; serine/threonine-protein kinase STK11; ubiquitin-like-conjugating enzyme ATG3; regulatory-associated protein of mTOR; autophagy protein 5; ras-related protein Rab-7a; 5’-AMP-activated protein kinase catalytic subunit alpha-2 |
| KEGG | KEGG:01100 | 0.0154985 | 0.1145620 | 1.156148 | 411796 551785 413867 550932 406081 552007 412876 551762 409624 409267 551968 413987 410980 412393 412541 413071 413663 550785 413233 411525 409884 409487 408470 551314 550804 550884 550767 551712 410096 408373 408868 726156 550667 725146 551721 725119 552755 408859 726310 411563 552421 409473 410798 409299 408930 551667 412273 727293 409329 552522 411188 551093 409515 408546 724361 552086 725623 551578 408461 413655 410627 410076 413340 551958 726747 406080 726239 724666 551447 551866 409270 552699 412362 408809 411959 413879 412632 552342 726095 409860 552496 408288 411692 552286 408441 408817 406114 413735 409155 412328 409250 411581 551182 412467 411897 409614 552533 551103 408969 409199 551964 409861 411698 551841 412426 100577378 413228 724239 413854 412245 727071 409846 409276 552180 724811 551102 727456 100577053 410059 726818 410105 551662 410396 552557 551593 412675 551837 412815 724436 406076 551775 408446 411447 100187709 412782 408991 411411 552023 | Metabolic pathways | ln | serine hydroxymethyltransferase; 6-phosphogluconolactonase; transaldolase; arginine kinase; glucose oxidase; pyruvate kinase; pyruvate carboxylase, mitochondrial; adenosylhomocysteinase 2-like; acetyl-coenzyme A synthetase; glycogen phosphorylase; aldose reductase-like; polypeptide N-acetylgalactosaminyltransferase 5; polyphosphoinositide phosphatase; cytosolic non-specific dipeptidase; long-chain-fatty-acid–CoA ligase 6; eye-specific diacylglycerol kinase; myotubularin-related protein 4; fructose-bisphosphate aldolase; methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial; choline/ethanolamine kinase; xylulose kinase; probable glutamine-dependent NAD(+) synthetase; NAD kinase-like; heparan-alpha-glucosaminide N-acetyltransferase-like; transketolase; N-sulphoglucosamine sulphohydrolase; ribose 5-phosphate isomerase A; lipoyltransferase 1, mitochondrial; gamma-glutamyltranspeptidase 1-like; phosphatidylinositol 4-kinase beta; inositol monophosphatase 2-like; pantothenate kinase 3; succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial; very-long-chain enoyl-CoA reductase; V-type proton ATPase subunit B; phospholipase D2; probable GDP-L-fucose synthase; pyrroline-5-carboxylate reductase 2; GPI ethanolamine phosphate transferase 1-like; myotubularin-related protein 2; glycogenin-1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; DNA methyltransferase 3; adenylosuccinate synthetase; tyrosine hydroxylase; bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial; probable chitinase 3; guanine deaminase; mannose-1-phosphate guanyltransferase beta; alpha-(1,6)-fucosyltransferase; L-lactate dehydrogenase-like; V-type proton ATPase catalytic subunit A; long-chain-fatty-acid–CoA ligase 4; phosphatidylinositide phosphatase SAC2; GPI inositol-deacylase; aldose 1-epimerase; heparanase-like; protein O-mannosyl-transferase 2; uridine 5’-monophosphate synthase; 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial; putative aminopeptidase W07G4.4; probable uridine-cytidine kinase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; uncharacterized LOC551958; cytochrome b-c1 complex subunit 8; polycomblike; putative lipoyltransferase 2, mitochondrial; 2-aminoethanethiol dioxygenase; mannose-6-phosphate isomerase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial; glycerol-3-phosphate acyltransferase 4; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; glucose 1,6-bisphosphate synthase; diacylglycerol kinase theta; fatty acid synthase-like; CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase, mitochondrial; adenylate kinase 1; lysophospholipid acyltransferase 2; galactoside 2-alpha-L-fucosyltransferase 2-like; GPI ethanolamine phosphate transferase 3; polypeptide N-acetylgalactosaminyltransferase 35A-like; isovaleryl-CoA dehydrogenase, mitochondrial; elongation of very long chain fatty acids protein 4-like; acetyl-CoA carboxylase; adenosine kinase 1; alanine–glyoxylate aminotransferase 2-like; alpha-amylase; glutamate–cysteine ligase regulatory subunit; dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial-like; NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial; beta-ureidopropionase; phosphopantothenate–cysteine ligase; phosphatidylinositol 4-kinase alpha; bifunctional purine biosynthesis protein PURH; phosphoglucomutase; group XIIA secretory phospholipase A2; GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase; probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial; aminoacylase-1-like; glycerol kinase; UDP-N-acetylhexosamine pyrophosphorylase; nucleoside diphosphate kinase; ethanolaminephosphotransferase 1-like; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; ubiquinone biosynthesis monooxygenase COQ6, mitochondrial; galactokinase-like; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; kynurenine/alpha-aminoadipate aminotransferase, mitochondrial-like; xylosyltransferase oxt; acidic mammalian chitinase; acylglycerol kinase, mitochondrial; GMP synthase [glutamine-hydrolyzing]; phosphatidylinositol 5-phosphate 4-kinase type-2 alpha; cob(I)yrinic acid a,c-diamide adenosyltransferase, mitochondrial-like; kynurenine–oxoglutarate transaminase 3-like; S-adenosylmethionine synthase; glucose-6-phosphate 1-epimerase; glycine dehydrogenase (decarboxylating), mitochondrial; probable citrate synthase 2, mitochondrial; beta-hexosaminidase subunit beta-like; alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase; beta-1,4-galactosyltransferase 7; isocitrate dehydrogenase [NAD] subunit gamma, mitochondrial; lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial; sphingosine-1-phosphate lyase; aspartate aminotransferase, mitochondrial; long-chain-fatty-acid–CoA ligase ACSBG2; fatty acid synthase; phospholipase A2-like; vacuolar H+ ATP synthase 16 kDa proteolipid subunit; trifunctional enzyme subunit beta, mitochondrial; probable aconitate hydratase, mitochondrial; serine palmitoyltransferase 2; nicotinamide riboside kinase; nucleoside diphosphate kinase 7; glutaminase kidney isoform, mitochondrial; NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial; guanylate kinase |
| KEGG | KEGG:00051 | 0.0188415 | 0.1145620 | 1.519598 | 551968 550785 552755 409329 551447 | Fructose and mannose metabolism | ln | aldose reductase-like; fructose-bisphosphate aldolase; probable GDP-L-fucose synthase; mannose-1-phosphate guanyltransferase beta; mannose-6-phosphate isomerase |
| KEGG | KEGG:00630 | 0.0253139 | 0.1145620 | 1.455054 | 411796 409624 | Glyoxylate and dicarboxylate metabolism | ln | serine hydroxymethyltransferase; acetyl-coenzyme A synthetase |
GO and KEGG enrichment for transcriptional modules
Here, we use hypergeometric tests on each of the 9 modules, testing for enrichment of GO terms in each module.
# Find the names for the genes in a given module
genes.in.module <- function(module.number){
colnames(OGGs[[1]])[network[[1]]$colors == paste("Module", module.number)]
}
# Get the gene names, and sensitivity to pheromone, for genes in a certain module
inspect.module.genes <- function(module){
gene.names <- tbl(my_db, "bee_names") %>% as.data.frame()
gene.names <- gene.names[gene.names$gene %in% genes.in.module(module), ]
gene.names$k <- colSums(as.matrix(TOM)[network[[1]]$colors == paste("Module", module),
network[[1]]$colors == paste("Module", module)])
gene.names <- gene.names %>% arrange(-k)
am <- tbl(my_db, "ebseq_gene_am") %>% as.data.frame()
gene.names$am_fc <- am$PostFC[match(gene.names$gene, am$gene)]
bt <- tbl(my_db, "ebseq_gene_bt") %>% rename(bt = gene) %>%
left_join(tbl(my_db, "bt2am")) %>% as.data.frame()
gene.names$bt_fc <- bt$PostFC[match(gene.names$gene, bt$am)]
lf <- tbl(my_db, "ebseq_gene_lf") %>% rename(lf = gene) %>%
left_join(tbl(my_db, "lf2am")) %>% as.data.frame()
gene.names$lf_fc <- lf$PostFC[match(gene.names$gene, lf$am)]
ln <- tbl(my_db, "ebseq_gene_ln") %>% rename(ln = gene) %>%
left_join(tbl(my_db, "ln2am")) %>% as.data.frame()
gene.names$ln_fc <- ln$PostFC[match(gene.names$gene, ln$am)]
row.names(gene.names) <- NULL
gene.names
}
GO.and.KEGG.hypergeometric <- function(gene_set, gene_universe, apis.db, min.size = 5, keep.all = FALSE){
p <- 0.05; if(keep.all) p <- 1
neatness <- function(x) format(round(x, 4), nsmall = 4) # for rounding
GO.enrichment <- function(gene_set, gene_universe, ontol){
result <- enrichGO(gene_set, apis.db, ont = ontol,
pvalueCutoff = p, universe = gene_universe,
qvalueCutoff = 1, minGSSize = min.size, maxGSSize = 500)
if(is.null(result)) return(NULL)
if(nrow(result@result) == 0) return(NULL)
result <- gofilter(result, level = 4) # Filter to high-level GO only
if(is.null(result)) return(NULL)
if(nrow(result@result) == 0) return(NULL)
result <- result@result
is_enriched <- sapply(result$GeneRatio, function(x) eval(parse(text=x))) > sapply(result$BgRatio, function(x) eval(parse(text=x)))
result <- result[is_enriched, ] %>% mutate(p.adjust = p.adjust(pvalue, method = "BH"))
Test_type <- "GO: Biological process"
if(ontol == "MF") Test_type <- "GO: Molecular function"
if(ontol == "CC") Test_type <- "GO: Cellular component"
data.frame(Test_type = Test_type, result, stringsAsFactors = FALSE)
}
kegg.enrichment <- function(gene_set, gene_universe){
result <- enrichKEGG(gene_set, organism = "ame", keyType = "kegg", pvalueCutoff = p,
gene_universe, minGSSize = min.size, maxGSSize = 500,
qvalueCutoff = 1, use_internal_data = FALSE, pAdjustMethod = "BH")
if(is.null(result)) return(NULL)
result <- result@result
is_enriched <- sapply(result$GeneRatio, function(x) eval(parse(text=x))) > sapply(result$BgRatio, function(x) eval(parse(text=x)))
result <- result[is_enriched, ] %>% mutate(p.adjust = p.adjust(pvalue, method = "BH"))
if(nrow(result %>% filter(pvalue < p)) == 0) return(NULL)
data.frame(Test_type = "KEGG", result, stringsAsFactors = FALSE)
}
rbind(kegg.enrichment(gene_set, gene_universe),
GO.enrichment(gene_set, gene_universe, "BP"),
GO.enrichment(gene_set, gene_universe, "MF"),
GO.enrichment(gene_set, gene_universe, "CC")) %>%
mutate(ID = str_replace_all(ID, "ame", "KEGG:"),
geneID = str_replace_all(geneID, "/", " "))
}
make_module_GO_table <- function(){
add_name_col <- function(df){
sapply(df$geneID, function(x){
data.frame(gene = x, stringsAsFactors = FALSE) %>%
left_join(tbl(my_db, "apis_entrez_names") %>% collect(), by = c("gene" = "entrez")) %>% .$name %>%
paste0(collapse = "; ")
}) -> df$gene_names
df %>% mutate(geneID = map_chr(geneID, function(x) paste0(x, collapse = " ")))
}
hub <- AnnotationHub::AnnotationHub()
select <- dplyr::select
rename <- dplyr::rename
filter <- dplyr::filter
apis.db <- hub[["AH62534"]]
gene.universe.modules <- entrez.tbl$entrez.id[entrez.tbl$gene %in% OGGs[[2]]$am] %>% as.character()
results <- lapply(1:9, function(i) { # or c(1,4,9)
gene_set <- entrez.tbl$entrez.id[entrez.tbl$gene %in% genes.in.module(i)] %>% as.character()
df <- GO.and.KEGG.hypergeometric(gene_set, gene.universe.modules, apis.db)
if(is.null(df) || nrow(df) == 0) return(NULL)
# # enrichment ratio: proportion genes in sample / proportion in gene universe
df$enrichment <- sapply(1:nrow(df), function(i) eval(parse(text = df$GeneRatio[i]))) /
sapply(1:nrow(df), function(i) eval(parse(text = df$BgRatio[i])))
df %>% mutate(Module = paste("Module", i)) %>% arrange(pvalue)
}) %>% do.call("rbind", .) %>%
mutate(Description = factor(Description, unique(Description))) %>%
filter(pvalue <= 0.05) # Only keep GO terms where the un-adjusted p is significant
results$sig <- "no" # Use a more strict classification, since the test has higher power
results$sig[results$p.adjust <= 0.05] <- "yes" # note: ADJUSTED p
results$sig <- relevel(factor(results$sig), ref = "yes")
results <- select(results, Module, Test_type, ID, Description, GeneRatio, BgRatio,
enrichment, pvalue, p.adjust, qvalue, Count, geneID, sig)
results$geneID <- strsplit(results$geneID, split = " ")
results %>% add_name_col()
}
module_enrichment_plot <- function(df, is.KEGG = FALSE){
label <- "Name of GO term"
if(is.KEGG) label <- "Name of KEGG term"
df %>% arrange(Module, log2(enrichment)) %>%
mutate(Description = factor(Description, unique(Description)),
Test_type = factor(Test_type, c("GO: Biological process", "GO: Molecular function", "GO: Cellular component", "KEGG"))) %>%
ggplot(aes(Description, log2(enrichment))) +
geom_bar(aes(fill = sig), stat = "identity", colour = "grey10") +
facet_wrap(~Module) +
scale_fill_brewer(palette = "Accent", direction = -1) +
scale_x_discrete(labels = function(x) str_wrap(x, width = 60)) +
theme_bw() +
theme(legend.position = "none", strip.background = element_blank(), strip.text = element_text(size = 10)) +
ylab("Log2 fold enrichment") + xlab(label) +
coord_flip()
}
module_GO_table <- make_module_GO_table()Figures of the module enrichment results
KEGG
module_KEGG <- module_GO_table %>% filter(Test_type == "KEGG", Module %in% c("Module 1", "Module 4", "Module 9")) %>%
module_enrichment_plot(is.KEGG = TRUE)
ggsave(module_KEGG, file = "figures/Figure 4 - module KEGG.pdf", height = 7, width = 8)
module_KEGG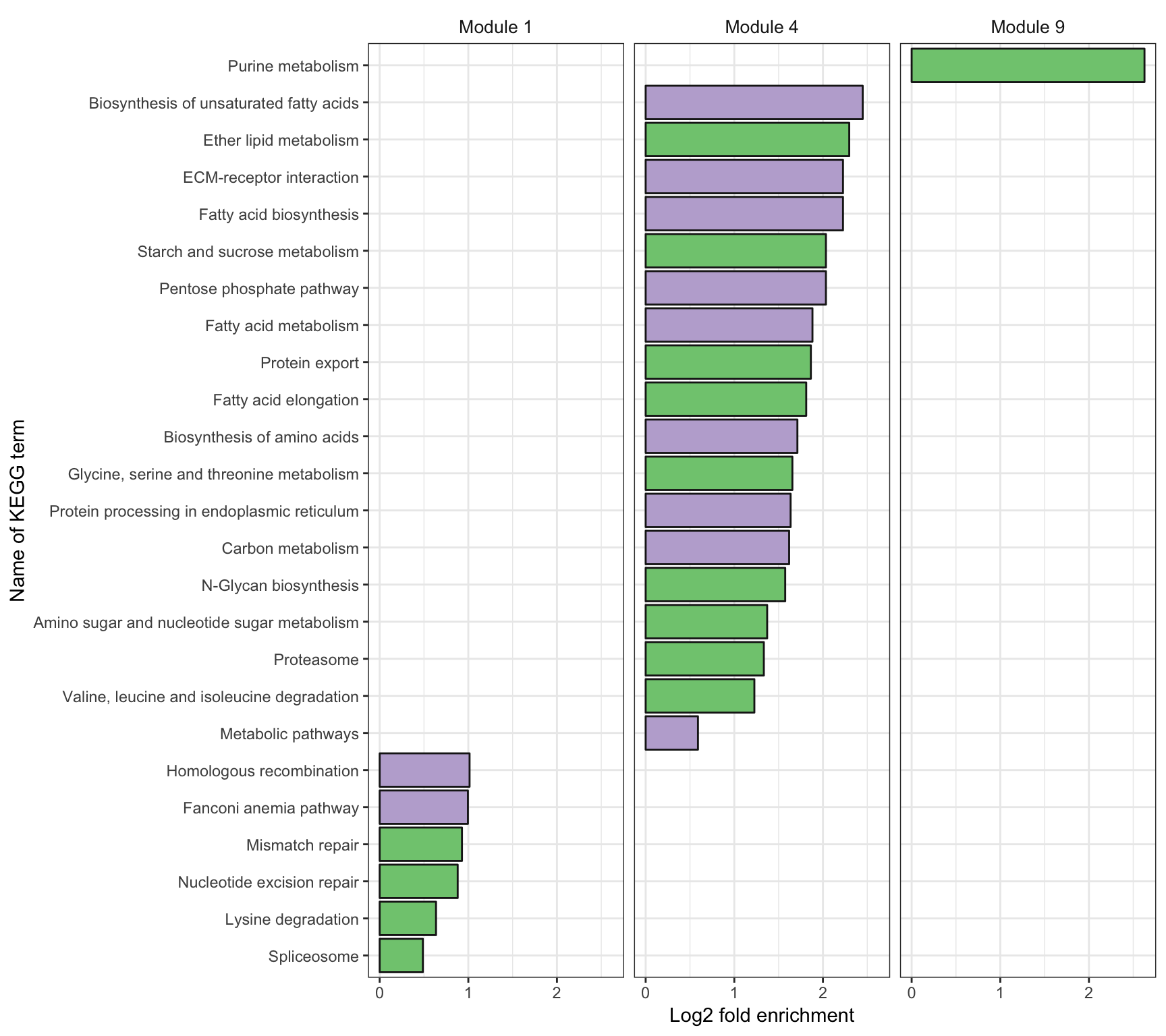
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Figure 4: Results of KEGG pathway enrichment analysis for the genes in each of the three significantly pheromone-sensitive transcriptional modules. The gene universe was defined as all genes for which we found an ortholog in all four species (i.e. the set that was used to discover these co-expressed modules). All KEGG terms shown in green were significantly enriched (p < 0.05), and those shown in purple remained significant after correction for multiple testing. Fold enrichment was calculated as the proportion of genes associated with the focal KEGG term in the module, divided by the equivalent proportion in the gene universe.
GO BP
fig_S6 <- module_GO_table %>%
filter(Test_type == "GO: Biological process", Module %in% c("Module 1", "Module 4", "Module 9")) %>%
module_enrichment_plot()
saveRDS(fig_S6, file = "supplement/fig_S6.rds")
fig_S6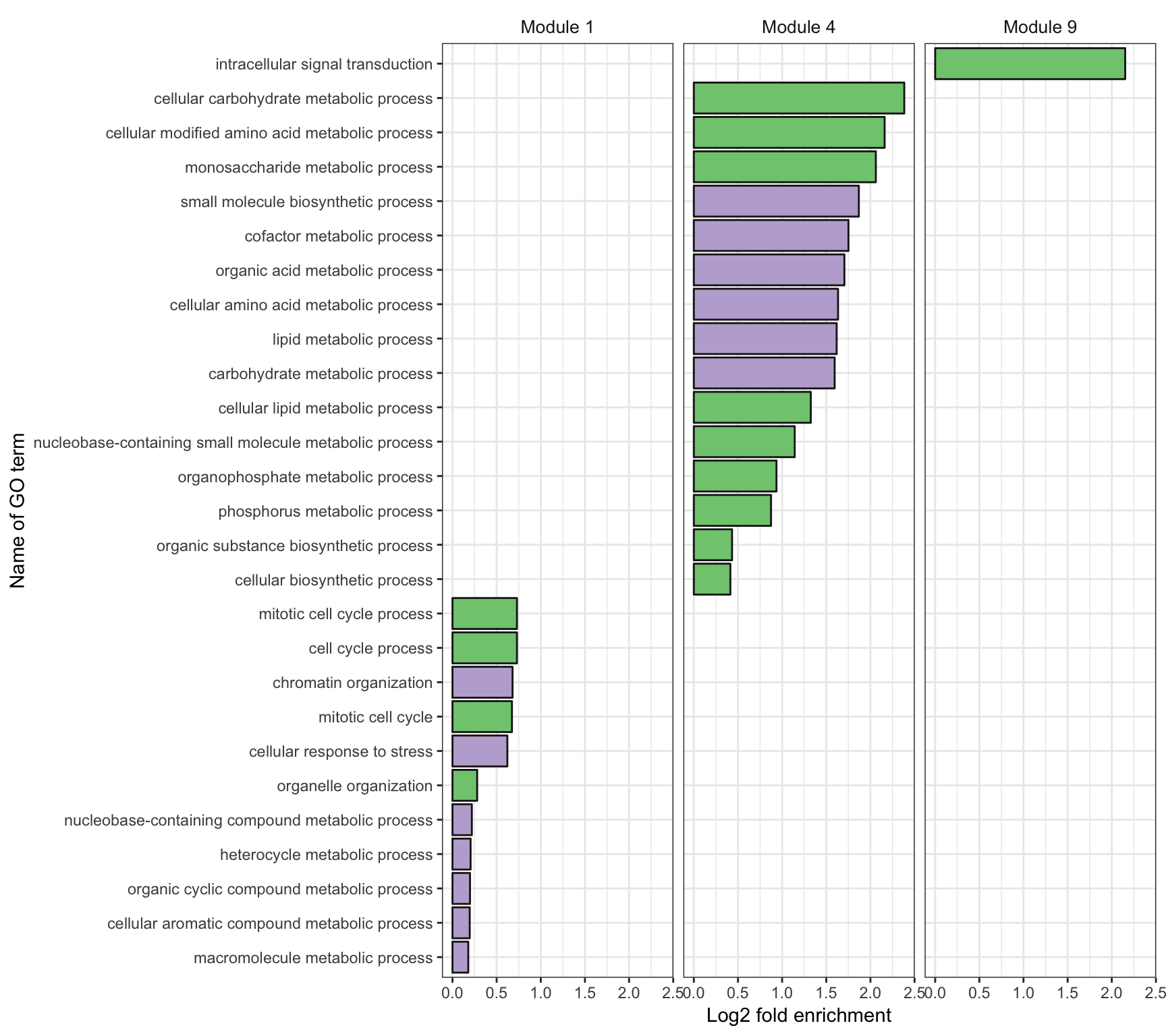
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Figure S6: Comparable figure to Figure 4, showing the results of GO: Biological process enrichment analysis instead of KEGG pathways.
GO MF
fig_S7 <- module_GO_table %>%
filter(Test_type == "GO: Molecular function", Module %in% c("Module 1", "Module 4", "Module 9")) %>%
module_enrichment_plot()
saveRDS(fig_S7, file = "supplement/fig_S7.rds")
fig_S7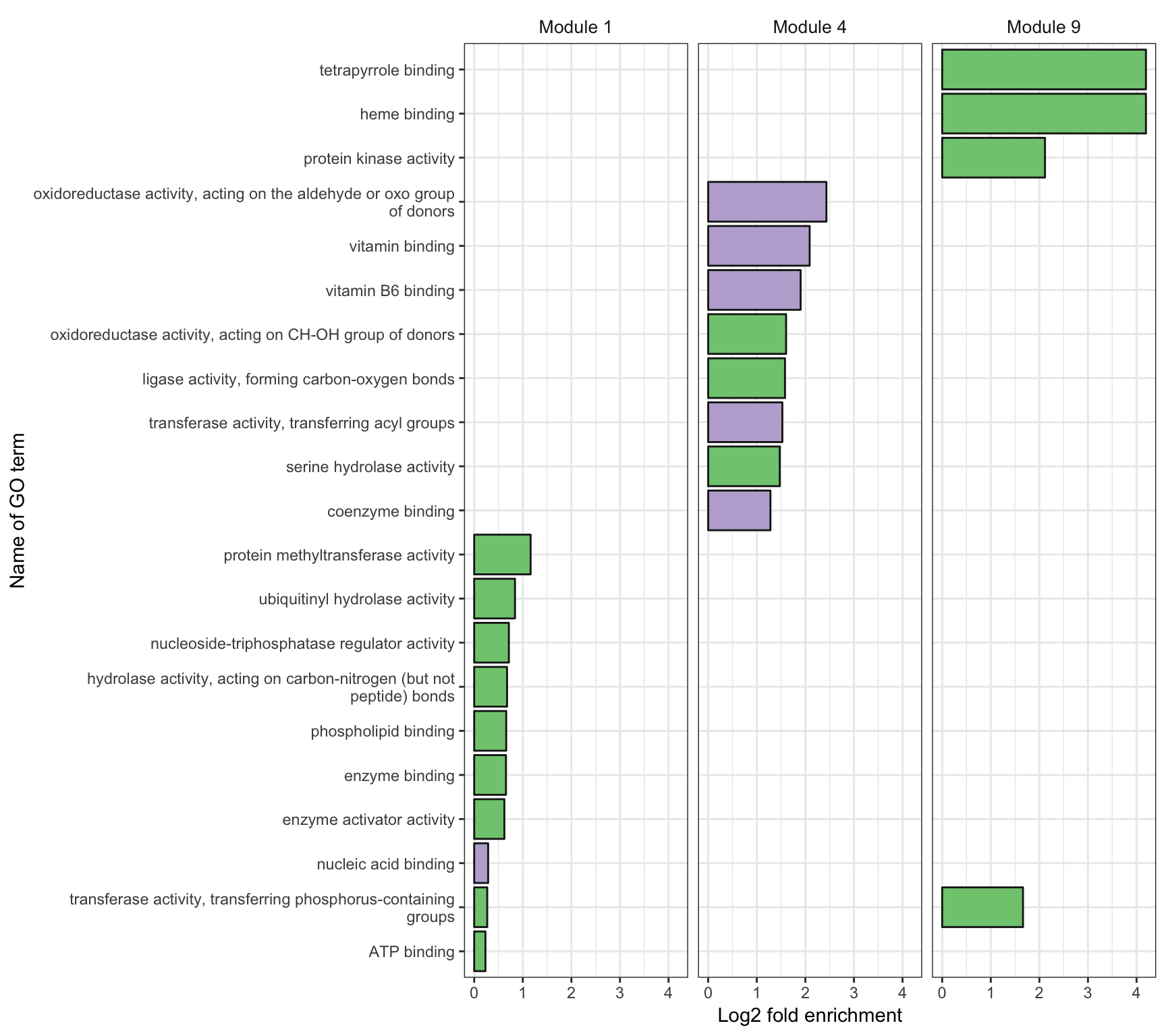
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Figure S7: Comparable figure to Figure 4, showing the results of GO: Molecular function enrichment analysis instead of KEGG pathways.
GO CC
fig_S8 <- module_GO_table %>%
filter(Test_type == "GO: Cellular component", Module %in% c("Module 1", "Module 4", "Module 9")) %>%
module_enrichment_plot()
saveRDS(fig_S8, file = "supplement/fig_S8.rds")
fig_S8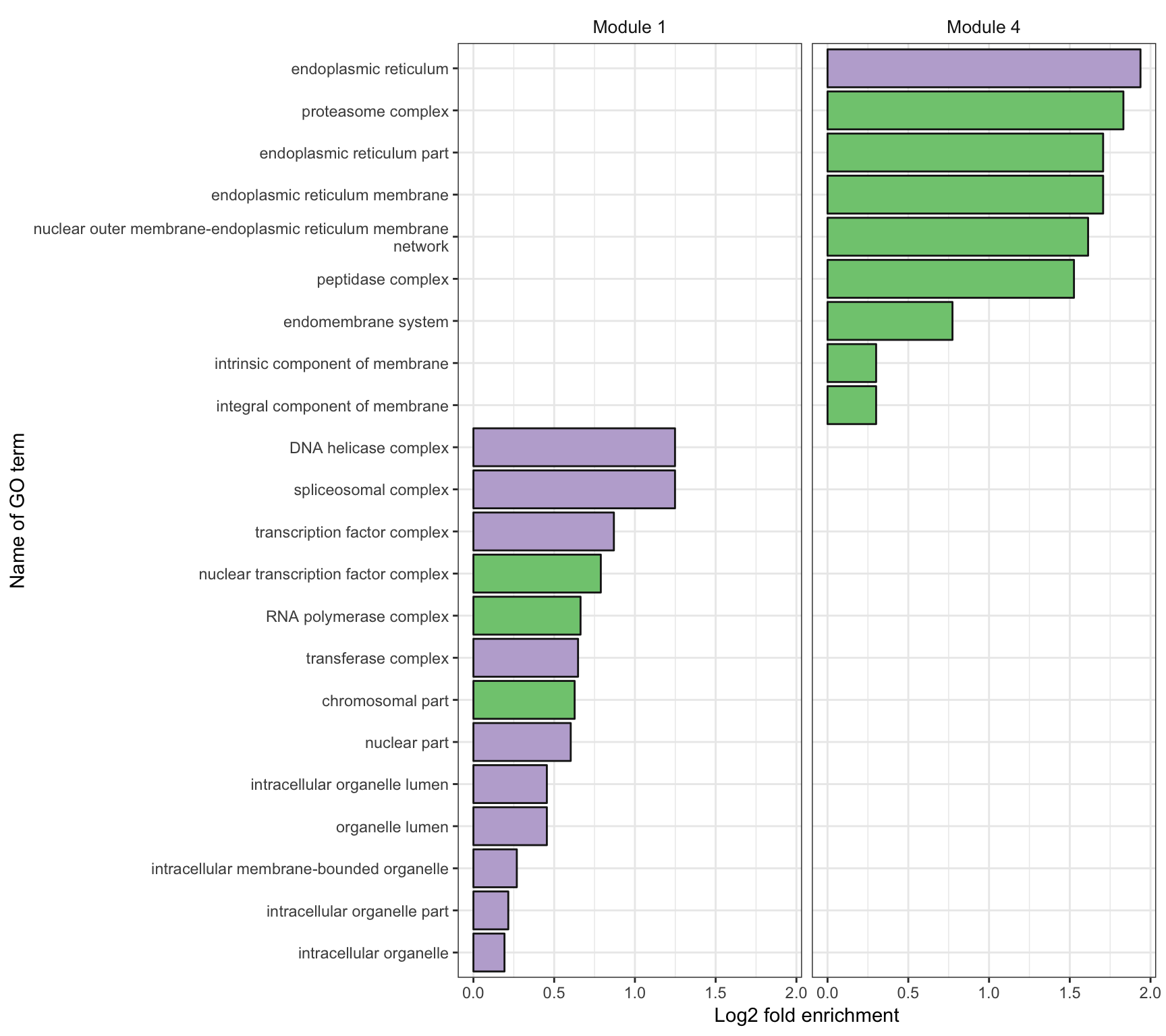
| Version | Author | Date |
|---|---|---|
| f260a62 | Luke Holman | 2019-03-07 |
Figure S8: Comparable figure to Figure 4, showing the results of GO: Cellular component enrichment analysis instead of KEGG pathways. Module 9 is missing because no GO:CC terms were significantly enriched.
Table of all module enrichment results
Table S24: List of every significant enrichment test result for each module, for all four ontologies. The latter two columns specify all the genes associated with the focal GO or KEGG term that are found in the module. The GeneRatio and BgRatio columns give the number of genes annotated with the focal term that are present in the focal module or the gene universe, respecitvely. These values were used to calculate the enrichment column, as the proportion of genes associated with the focal annotation term in the module, divided by the equivalent proportion in the gene universe.
tab_S24 <- module_GO_table %>% select(-sig, -Count)
saveRDS(tab_S24 %>%
select(-geneID, -gene_names, -qvalue, -GeneRatio, -BgRatio) %>%
mutate(Test_type = str_replace_all(Test_type, " Biological process", "BP"),
Test_type = str_replace_all(Test_type, " Molecular function", "MF"),
Test_type = str_replace_all(Test_type, " Cellular component", "CC")), file = "supplement/tab_S24.rds")
kable.table(tab_S24)| Module | Test_type | ID | Description | GeneRatio | BgRatio | enrichment | pvalue | p.adjust | qvalue | geneID | gene_names |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Module 1 | GO: Cellular component | GO:0044428 | nuclear part | 62/474 | 97/1126 | 1.518378 | 0.0000050 | 0.0001739 | 0.0003165 | 408606 408611 408615 408632 409049 409423 409544 409735 409810 409906 409994 410195 410200 410343 410395 410459 410855 410864 410907 411072 411265 411279 411433 411465 411619 411918 411985 412072 412077 412199 412268 412377 412589 413181 413351 413404 413499 413523 413548 413793 413963 550895 550924 551131 551383 551470 551620 551745 551974 552353 552449 552563 552601 552696 552703 552780 552794 724855 725905 726816 100576104 100578879 | PRP3 pre-mRNA processing factor 3 homolog; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; spliceosome-associated protein CWC15 homolog; paired amphipathic helix protein Sin3a; male-specific lethal 1 homolog; ruvB-like 1; transcription initiation factor TFIID subunit 6; anaphase-promoting complex subunit 4; transcription initiation factor TFIID subunit 2; INO80 complex subunit E; COP9 signalosome complex subunit 6; mRNA turnover protein 4 homolog; nuclear pore complex protein Nup93-like; chromobox protein homolog 1-like; cyclin-H; retinoblastoma-binding protein 5 homolog; nuclear pore complex protein Nup50; U4/U6 small nuclear ribonucleoprotein Prp31; cysteine-rich protein 2-binding protein-like; CXXC-type zinc finger protein 1-like; actin-related protein 8; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; double-strand-break repair protein rad21 homolog; nuclear pore complex protein Nup205; histone-lysine N-methyltransferase SETD1; DNA replication complex GINS protein PSF1-like; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; transcriptional adapter 1-like; transcription initiation factor IIA subunit 1; parafibromin; BRCA1-A complex subunit BRE-like; mortality factor 4-like protein 1; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; cell division cycle protein 23 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; enhancer of polycomb homolog 1; splicing factor 3A subunit 3; general transcription factor IIH subunit 1; cleavage stimulation factor subunit 2; protein max; protein MAK16 homolog A; general transcription factor IIF subunit 2; pre-mRNA-processing-splicing factor 8; integrator complex subunit 10; thioredoxin-like protein 4A; DNA-directed RNA polymerase III subunit RPC4; mediator of RNA polymerase II transcription subunit 6; transcription initiation factor TFIID subunit 7; lys-63-specific deubiquitinase BRCC36-like; histone acetyltransferase KAT8; KAT8 regulatory NSL complex subunit 2; ribonuclease P/MRP protein subunit POP5; mediator of RNA polymerase II transcription subunit 14; BRISC and BRCA1-A complex member 1-like; pre-mRNA-splicing factor 18; ruvB-like 2; INO80 complex subunit B; protein CASC3 |
| Module 1 | GO: Molecular function | GO:0003676 | nucleic acid binding | 174/626 | 319/1400 | 1.219866 | 0.0000397 | 0.0014682 | 0.0082704 | 406077 406084 406123 408295 408336 408352 408386 408580 408611 408615 408620 408710 408711 408724 408815 409028 409059 409098 409119 409251 409259 409340 409392 409493 409550 409557 409573 409696 409833 409839 409866 409883 409887 409915 410027 410203 410326 410328 410376 410410 410434 410571 410749 410757 410876 410918 411103 411126 411164 411265 411293 411351 411407 411450 411492 411538 411640 411649 411705 411780 411786 411833 411854 411985 412077 412190 412410 412491 412574 412750 413055 413087 413091 413139 413146 413176 413498 413558 413650 413690 413794 413815 413963 414004 550692 550858 550924 551083 551240 551242 551309 551398 551470 551538 551540 551586 551602 551616 551620 551739 551825 551840 551997 552022 552027 552056 552072 552112 552151 552172 552206 552247 552255 552303 552308 552336 552353 552450 552484 552509 552527 552539 552567 552768 552772 552787 552815 724150 724247 724296 724315 724363 724424 724810 724955 725012 725061 725189 725201 725220 725254 725268 725303 725330 725350 725441 725496 725515 725719 726058 726204 726385 726583 727087 727113 727284 727473 100576131 100576321 100576600 100576768 100576775 100576784 100577033 100577174 100577272 100577491 100578252 100578406 100578529 100578697 100578743 100578879 100578927 | homeotic protein antennapedia; ecdysone receptor; pipsqueak; probable serine/threonine-protein kinase DDB_G0282963; exonuclease mut-7 homolog; elongation factor Ts, mitochondrial; eukaryotic translation initiation factor 3 subunit G; eukaryotic translation initiation factor 2D; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; RNA-binding protein 42; KH domain-containing, RNA-binding, signal transduction-associated protein 2-like; zinc finger matrin-type protein 5-like; uncharacterized LOC408724; probable DNA mismatch repair protein Msh6; probable ATP-dependent RNA helicase YTHDC2; rabenosyn-5; histone-lysine N-methyltransferase eggless; REST corepressor 3; BRCA1-associated protein; protein abrupt; UV excision repair protein RAD23 homolog B; ATPase WRNIP1-like; zinc finger protein 569-like; eukaryotic translation initiation factor 3 subunit D; protein maelstrom; zinc finger protein 2 homolog; splicing factor 45; hemK methyltransferase family member 1; regulator of nonsense transcripts 1; eukaryotic initiation factor 4A-III; splicing factor 3A subunit 1; transcription factor Dp-1; RNA-binding protein 26; survival of motor neuron-related-splicing factor 30; POU domain protein CF1A; Krueppel-like factor 10; zinc finger protein 341-like; peptidyl-tRNA hydrolase ICT1, mitochondrial; ATP-dependent DNA helicase PIF1; splicing factor 3B subunit 4; DNA topoisomerase 3-alpha; squamous cell carcinoma antigen recognized by T-cells 3; circadian locomoter output cycles protein kaput; THUMP domain-containing protein 3-like; protein tramtrack, beta isoform; putative 28S ribosomal protein S5, mitochondrial; broad-complex core protein isoforms 1/2/3/4/5; DNA-directed RNA polymerase III subunit RPC3; CXXC-type zinc finger protein 1-like; zinc finger protein 622; elongation factor G, mitochondrial; leucine-rich repeat-containing protein 47-like; probable ATP-dependent RNA helicase DDX43; ATP-dependent RNA helicase DHX36; pre-mRNA-splicing factor RBM22; DNA replication licensing factor Mcm2; alkylated DNA repair protein alkB homolog 8; synaptojanin-1; zinc finger protein 578-like; general transcription factor IIE subunit 1; la protein homolog; heat shock factor protein; histone-lysine N-methyltransferase SETD1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; probable ATP-dependent RNA helicase DDX47; U3 small nucleolar RNA-associated protein 6 homolog; double-stranded RNA-specific editase 1-like; PR domain zinc finger protein 10-like; DNA topoisomerase 1; uncharacterized LOC413055; tRNA (uracil-5-)-methyltransferase homolog A-like; serrate RNA effector molecule homolog; probable phenylalanine–tRNA ligase, mitochondrial; regulator of nonsense transcripts 2; reticulocyte-binding protein 2 homolog a-like; F-box only protein 21-like; photoreceptor-specific nuclear receptor; nuclear RNA export factor 1-like; helicase SKI2W; translin; CCA tRNA nucleotidyltransferase 1, mitochondrial-like; splicing factor 3A subunit 3; eukaryotic translation initiation factor 3 subunit A; TATA-box-binding protein; RNA-binding protein 40; cleavage stimulation factor subunit 2; probable tRNA pseudouridine synthase 2; uncharacterized LOC551240; eukaryotic translation initiation factor 3 subunit E; transcription initiation factor TFIID subunit 1; DNA polymerase delta catalytic subunit; general transcription factor IIF subunit 2; lysine–tRNA ligase; replication factor C subunit 2; programmed cell death protein 5; ras GTPase-activating protein-binding protein 2; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1; pre-mRNA-processing-splicing factor 8; DNA primase large subunit; nuclear hormone receptor HR96; YTH domain-containing family protein 1; cleavage and polyadenylation specificity factor subunit 1; zinc finger CCCH-type with G patch domain-containing protein-like; heterogeneous nuclear ribonucleoprotein A1, A2/B1 homolog; eukaryotic translation initiation factor 3 subunit L; exosome complex component CSL4; RNA-binding protein 28; dnaJ homolog subfamily C member 1-like; G patch domain-containing protein 1 homolog; putative ATP-dependent RNA helicase me31b; nuclear factor NF-kappa-B p100 subunit; broad-complex; RNA-binding protein cabeza-like; zinc finger CCHC-type and RNA-binding motif-containing protein 1-like; UBX domain-containing protein 1; DNA-directed RNA polymerase III subunit RPC4; DNA repair protein RAD51 homolog A; uncharacterized LOC552484; MKI67 FHA domain-interacting nucleolar phosphoprotein-like; cell division cycle 5-like protein; density-regulated protein homolog; DEAD-box helicase Dbp80; phenylalanine–tRNA ligase alpha subunit; protein-lysine N-methyltransferase N6AMT2; DNA topoisomerase 3-beta-1; high mobility group protein 20A-like; B-cell lymphoma/leukemia 11B; rRNA methyltransferase 3, mitochondrial; DNA mismatch repair protein Mlh1; forkhead box protein D3-like; activator of basal transcription 1-like; sprT-like domain-containing protein Spartan; mucin-5AC; KH domain-containing protein akap-1; zinc finger protein 25-like; uncharacterized LOC725061; protein bric-a-brac 1-like; 39S ribosomal protein L21, mitochondrial; homeobox protein Nkx-6.1-like; endothelial zinc finger protein induced by tumor necrosis factor alpha; S1 RNA-binding domain-containing protein 1; histone H4 transcription factor; DNA polymerase delta small subunit; GATA-binding factor A; probable ATP-dependent RNA helicase DDX49; dimethyladenosine transferase 1, mitochondrial; zinc finger protein 543-like; DNA-directed RNA polymerase I subunit RPA2; FACT complex subunit Ssrp1; hairy/enhancer-of-split related with YRPW motif protein 1; GTPase Era, mitochondrial; DNA polymerase eta; serine/arginine-rich splicing factor 7; probable ATP-dependent RNA helicase spindle-E; transcription factor Sox-10-like; signal recognition particle 19 kDa protein; zinc finger protein 267-like; uncharacterized LOC100576321; zinc finger protein 345-like; nuclear cap-binding protein subunit 3-like; nucleolar MIF4G domain-containing protein 1 homolog; high mobility group B protein 6-like; zinc finger protein 33B; ras-responsive element-binding protein 1-like; uncharacterized LOC100577272; survival motor neuron protein; Ets at 97D ortholog; asparagine-rich zinc finger protein AZF1-like; zinc finger protein 701-like; THUMP domain-containing protein 1 homolog; zinc finger protein 431-like; protein CASC3; transcriptional repressor protein YY1-like |
| Module 1 | GO: Cellular component | GO:0043231 | intracellular membrane-bounded organelle | 178/474 | 351/1126 | 1.204683 | 0.0000556 | 0.0009725 | 0.0017696 | 406077 406084 408352 408493 408511 408548 408606 408611 408615 408632 408711 408808 408837 409049 409098 409119 409224 409331 409340 409423 409544 409586 409735 409765 409810 409887 409906 409994 410027 410195 410200 410203 410217 410298 410343 410387 410390 410395 410410 410459 410757 410855 410856 410864 410907 411044 411072 411265 411279 411304 411327 411351 411433 411465 411603 411619 411640 411654 411799 411833 411854 411865 411918 411985 412016 412072 412077 412199 412268 412278 412350 412377 412409 412506 412589 412710 412796 412984 413055 413078 413181 413271 413340 413351 413404 413490 413499 413523 413548 413558 413650 413666 413688 413793 413878 413940 413963 550716 550895 550924 551090 551131 551240 551383 551398 551427 551470 551620 551642 551692 551745 551781 551811 551825 551843 551961 551974 551997 552052 552167 552247 552253 552312 552353 552440 552449 552450 552484 552495 552522 552533 552563 552601 552696 552703 552763 552780 552794 552815 685996 724152 724205 724315 724559 724665 724855 724955 725012 725023 725061 725144 725220 725254 725515 725565 725719 725854 725905 726007 726058 726137 726204 726239 726617 726731 726816 727263 727284 100576104 100576131 100576600 100576667 100576784 100577491 100578252 100578450 100578879 100579040 | homeotic protein antennapedia; ecdysone receptor; elongation factor Ts, mitochondrial; uncharacterized LOC408493; protein preli-like; MICOS complex subunit Mic60; PRP3 pre-mRNA processing factor 3 homolog; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; spliceosome-associated protein CWC15 homolog; zinc finger matrin-type protein 5-like; prosaposin; cytochrome c oxidase subunit 5A, mitochondrial; paired amphipathic helix protein Sin3a; histone-lysine N-methyltransferase eggless; REST corepressor 3; ubiquinone biosynthesis protein COQ7; YEATS domain-containing protein 2; UV excision repair protein RAD23 homolog B; male-specific lethal 1 homolog; ruvB-like 1; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9; transcription initiation factor TFIID subunit 6; actin-related protein 6; anaphase-promoting complex subunit 4; transcription factor Dp-1; transcription initiation factor TFIID subunit 2; INO80 complex subunit E; survival of motor neuron-related-splicing factor 30; COP9 signalosome complex subunit 6; mRNA turnover protein 4 homolog; POU domain protein CF1A; structural maintenance of chromosomes protein 3; conserved oligomeric Golgi complex subunit 6; nuclear pore complex protein Nup93-like; conserved oligomeric Golgi complex subunit 3; protein suppressor of forked; chromobox protein homolog 1-like; ATP-dependent DNA helicase PIF1; cyclin-H; circadian locomoter output cycles protein kaput; retinoblastoma-binding protein 5 homolog; 28S ribosomal protein S29, mitochondrial; nuclear pore complex protein Nup50; U4/U6 small nuclear ribonucleoprotein Prp31; inhibitor of growth protein 1; cysteine-rich protein 2-binding protein-like; CXXC-type zinc finger protein 1-like; actin-related protein 8; reticulon-4-interacting protein 1, mitochondrial-like; G1/S-specific cyclin-E1; elongation factor G, mitochondrial; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; digestive organ expansion factor homolog; double-strand-break repair protein rad21 homolog; DNA replication licensing factor Mcm2; probable tRNA N6-adenosine threonylcarbamoyltransferase; N-acetylgalactosaminyltransferase 7; la protein homolog; heat shock factor protein; exportin-2; nuclear pore complex protein Nup205; histone-lysine N-methyltransferase SETD1; transforming growth factor beta regulator 1; DNA replication complex GINS protein PSF1-like; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; transcriptional adapter 1-like; transcription initiation factor IIA subunit 1; tuberin; histone deacetylase 3; parafibromin; mitochondrial import inner membrane translocase subunit TIM44; general vesicular transport factor p115; BRCA1-A complex subunit BRE-like; trafficking protein particle complex subunit 3; 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial; 28S ribosomal protein S30, mitochondrial; uncharacterized LOC413055; very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase; mortality factor 4-like protein 1; galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase I; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; clavesin-2-like; cell division cycle protein 23 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; photoreceptor-specific nuclear receptor; nuclear RNA export factor 1-like; peroxisome biogenesis factor 1; DNA-directed RNA polymerase III subunit RPC5; enhancer of polycomb homolog 1; glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial; importin-9; splicing factor 3A subunit 3; clathrin heavy chain; general transcription factor IIH subunit 1; cleavage stimulation factor subunit 2; conserved oligomeric Golgi complex subunit 8; protein max; uncharacterized LOC551240; protein MAK16 homolog A; DNA polymerase delta catalytic subunit; erlin-1-like; general transcription factor IIF subunit 2; pre-mRNA-processing-splicing factor 8; GPI mannosyltransferase 4; protein FAM50 homolog; integrator complex subunit 10; pre-rRNA-processing protein TSR1 homolog; graves disease carrier protein homolog; nuclear hormone receptor HR96; LETM1 and EF-hand domain-containing protein anon-60Da, mitochondrial; V-type proton ATPase subunit G; thioredoxin-like protein 4A; cleavage and polyadenylation specificity factor subunit 1; zinc finger protein 330 homolog; IWS1-like protein; nuclear factor NF-kappa-B p100 subunit; mitochondrial chaperone BCS1; RAD50-interacting protein 1; DNA-directed RNA polymerase III subunit RPC4; histone chaperone asf1; mediator of RNA polymerase II transcription subunit 6; DNA repair protein RAD51 homolog A; uncharacterized LOC552484; ATP-binding cassette sub-family D member 3; alpha-(1,6)-fucosyltransferase; GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase; transcription initiation factor TFIID subunit 7; lys-63-specific deubiquitinase BRCC36-like; histone acetyltransferase KAT8; KAT8 regulatory NSL complex subunit 2; cell division control protein 6 homolog; ribonuclease P/MRP protein subunit POP5; mediator of RNA polymerase II transcription subunit 14; high mobility group protein 20A-like; suppressor of variegation 3-9; uncharacterized LOC724152; protein AF-9; forkhead box protein D3-like; cell cycle checkpoint protein RAD17; protein SET; BRISC and BRCA1-A complex member 1-like; KH domain-containing protein akap-1; zinc finger protein 25-like; mediator of RNA polymerase II transcription subunit 26; uncharacterized LOC725061; uncharacterized LOC725144; homeobox protein Nkx-6.1-like; endothelial zinc finger protein induced by tumor necrosis factor alpha; zinc finger protein 543-like; nitric oxide synthase-interacting protein homolog; DNA-directed RNA polymerase I subunit RPA2; 39S ribosomal protein L37, mitochondrial; pre-mRNA-splicing factor 18; polycomb protein Scm; FACT complex subunit Ssrp1; male-specific lethal 3 homolog; hairy/enhancer-of-split related with YRPW motif protein 1; putative lipoyltransferase 2, mitochondrial; 39S ribosomal protein L48, mitochondrial; probable 28S ribosomal protein S26, mitochondrial; ruvB-like 2; chromobox protein homolog 5; transcription factor Sox-10-like; INO80 complex subunit B; zinc finger protein 267-like; zinc finger protein 345-like; E3 ubiquitin-protein ligase synoviolin A; high mobility group B protein 6-like; survival motor neuron protein; Ets at 97D ortholog; histone-lysine N-methyltransferase SETD2; protein CASC3; chondroitin sulfate synthase 2 |
| Module 1 | KEGG | KEGG:03440 | Homologous recombination | 13/497 | 14/1081 | 2.019690 | 0.0003052 | 0.0125127 | 0.0305187 | 409447 410571 412589 551398 552450 552601 552787 552851 724855 725330 725418 725934 726642 | DNA repair and recombination protein RAD54-like; DNA topoisomerase 3-alpha; BRCA1-A complex subunit BRE-like; DNA polymerase delta catalytic subunit; DNA repair protein RAD51 homolog A; lys-63-specific deubiquitinase BRCC36-like; DNA topoisomerase 3-beta-1; crossover junction endonuclease EME1; BRISC and BRCA1-A complex member 1-like; DNA polymerase delta small subunit; replication protein A 32 kDa subunit; replication protein A 70 kDa DNA-binding subunit; DNA repair protein RAD50 |
| Module 1 | GO: Biological process | GO:0043170 | macromolecule metabolic process | 226/463 | 414/960 | 1.131875 | 0.0003744 | 0.0228373 | 0.0298192 | 406084 406112 408346 408386 408501 408527 408606 408611 408615 408632 408675 408687 408724 408815 408924 408980 409049 409140 409191 409198 409279 409331 409340 409347 409387 409389 409392 409394 409423 409479 409544 409550 409557 409637 409653 409696 409723 409735 409810 409839 409860 409883 409887 409891 409915 410027 410093 410162 410195 410203 410390 410395 410410 410459 410571 410749 410757 410806 410907 410941 410958 411044 411103 411164 411167 411218 411273 411279 411351 411362 411433 411465 411572 411640 411649 411654 411719 411786 411799 411833 411981 411985 412072 412159 412200 412219 412268 412350 412377 412410 412484 412580 412644 412676 412727 412750 412868 412984 413087 413139 413181 413271 413351 413404 413523 413548 413558 413643 413650 413688 413690 413718 413738 413793 413794 413813 413815 413821 413878 413963 414001 414004 550692 550698 550895 550924 551083 551150 551158 551240 551242 551256 551321 551343 551358 551386 551398 551427 551470 551497 551538 551540 551578 551616 551620 551745 551771 551812 551825 551855 551972 551974 552056 552172 552178 552186 552303 552324 552353 552449 552450 552458 552484 552522 552529 552554 552563 552593 552601 552660 552676 552696 552733 552763 552768 552780 552787 724152 724186 724205 724244 724247 724296 724338 724424 724467 724496 724559 724596 724635 724708 724855 724879 725054 725158 725201 725220 725330 725592 725633 725719 725813 725854 725859 725905 725948 726007 726058 726086 726137 726151 726204 726239 726449 726583 726816 727192 100576667 100576866 100577386 100577491 100578201 100578266 100578450 100578852 100578879 | ecdysone receptor; period circadian protein; UPF0396 protein CG6066; eukaryotic translation initiation factor 3 subunit G; serine/threonine-protein kinase/endoribonuclease IRE1; pre-mRNA-splicing factor SPF27; PRP3 pre-mRNA processing factor 3 homolog; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; spliceosome-associated protein CWC15 homolog; probable 39S ribosomal protein L24, mitochondrial; putative tRNA (cytidine(32)/guanosine(34)-2’-O)-methyltransferase; uncharacterized LOC408724; probable DNA mismatch repair protein Msh6; peptidoglycan-recognition protein LC; DNA oxidative demethylase ALKBH1; paired amphipathic helix protein Sin3a; transcription elongation factor B polypeptide 1; SUMO-activating enzyme subunit 2; 26S protease regulatory subunit 6A-B; cullin-4A; YEATS domain-containing protein 2; UV excision repair protein RAD23 homolog B; U4/U6.U5 tri-snRNP-associated protein 1; ubiquitin carboxyl-terminal hydrolase; ubiquitin specific protease-like; ATPase WRNIP1-like; NEDD8-activating enzyme E1 catalytic subunit; male-specific lethal 1 homolog; 60S ribosomal protein L6; ruvB-like 1; eukaryotic translation initiation factor 3 subunit D; protein maelstrom; 60S ribosomal protein L23a; tRNA-splicing ligase RtcB homolog; splicing factor 45; 40S ribosomal protein S12, mitochondrial; transcription initiation factor TFIID subunit 6; anaphase-promoting complex subunit 4; regulator of nonsense transcripts 1; GPI ethanolamine phosphate transferase 3; splicing factor 3A subunit 1; transcription factor Dp-1; cell division cycle and apoptosis regulator protein 1; RNA-binding protein 26; survival of motor neuron-related-splicing factor 30; geminin; ataxin-3-like; COP9 signalosome complex subunit 6; POU domain protein CF1A; protein suppressor of forked; chromobox protein homolog 1-like; ATP-dependent DNA helicase PIF1; cyclin-H; DNA topoisomerase 3-alpha; squamous cell carcinoma antigen recognized by T-cells 3; circadian locomoter output cycles protein kaput; histidine–tRNA ligase, cytoplasmic; U4/U6 small nuclear ribonucleoprotein Prp31; protein MTO1 homolog, mitochondrial; ubiquitin-like modifier-activating enzyme 1; inhibitor of growth protein 1; putative 28S ribosomal protein S5, mitochondrial; DNA-directed RNA polymerase III subunit RPC3; cysteine–tRNA ligase, cytoplasmic; WW domain-binding protein 11; uncharacterized protein DDB_G0287625-like; actin-related protein 8; elongation factor G, mitochondrial; ubiquitin carboxyl-terminal hydrolase 8-like; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; ubiquitin carboxyl-terminal hydrolase 14; DNA replication licensing factor Mcm2; alkylated DNA repair protein alkB homolog 8; probable tRNA N6-adenosine threonylcarbamoyltransferase; probable queuine tRNA-ribosyltransferase; general transcription factor IIE subunit 1; N-acetylgalactosaminyltransferase 7; la protein homolog; probable ubiquitin carboxyl-terminal hydrolase FAF-X; histone-lysine N-methyltransferase SETD1; DNA replication complex GINS protein PSF1-like; pre-mRNA-splicing factor SYF1; threonine–tRNA ligase, cytoplasmic; ubiquitin conjugation factor E4 B; transcription initiation factor IIA subunit 1; histone deacetylase 3; parafibromin; U3 small nucleolar RNA-associated protein 6 homolog; peptidoglycan recognition protein S2; protein arginine N-methyltransferase 3; josephin-2; protein mahjong; cytosolic carboxypeptidase-like protein 5; DNA topoisomerase 1; ubiquitin fusion degradation protein 1 homolog; 28S ribosomal protein S30, mitochondrial; tRNA (uracil-5-)-methyltransferase homolog A-like; probable phenylalanine–tRNA ligase, mitochondrial; mortality factor 4-like protein 1; galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase I; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; photoreceptor-specific nuclear receptor; N-acetylglucosaminyl-phosphatidylinositol biosynthetic protein; nuclear RNA export factor 1-like; DNA-directed RNA polymerase III subunit RPC5; helicase SKI2W; probable glutamine–tRNA ligase; uncharacterized LOC413738; enhancer of polycomb homolog 1; translin; ubiquitin carboxyl-terminal hydrolase isozyme L5; CCA tRNA nucleotidyltransferase 1, mitochondrial-like; histone deacetylase complex subunit SAP130-A-like; glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial; splicing factor 3A subunit 3; mRNA-capping enzyme; eukaryotic translation initiation factor 3 subunit A; TATA-box-binding protein; cyclin-Y; general transcription factor IIH subunit 1; cleavage stimulation factor subunit 2; probable tRNA pseudouridine synthase 2; BRCA2-interacting transcriptional repressor EMSY; 28S ribosomal protein S17, mitochondrial; uncharacterized LOC551240; eukaryotic translation initiation factor 3 subunit E; protein CNPPD1; 39S ribosomal protein L44, mitochondrial; 26S protease regulatory subunit 7; dehydrodolichyl diphosphate syntase complex subunit DHDDS; 26S protease regulatory subunit 10B; DNA polymerase delta catalytic subunit; erlin-1-like; general transcription factor IIF subunit 2; lariat debranching enzyme; lysine–tRNA ligase; replication factor C subunit 2; protein O-mannosyl-transferase 2; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1; pre-mRNA-processing-splicing factor 8; integrator complex subunit 10; methionine aminopeptidase 2; mitochondrial tRNA-specific 2-thiouridylase 1; nuclear hormone receptor HR96; exosome complex component rrp45-like; alpha-1,3/1,6-mannosyltransferase ALG2; thioredoxin-like protein 4A; eukaryotic translation initiation factor 3 subunit L; G patch domain-containing protein 1 homolog; nicastrin; cell division cycle protein 20 homolog; RNA-binding protein cabeza-like; ubiquitin carboxyl-terminal hydrolase 5; DNA-directed RNA polymerase III subunit RPC4; mediator of RNA polymerase II transcription subunit 6; DNA repair protein RAD51 homolog A; DNA-directed RNA polymerase II subunit RPB3; uncharacterized LOC552484; alpha-(1,6)-fucosyltransferase; U6 snRNA-associated Sm-like protein LSm4; RNA 3’-terminal phosphate cyclase-like; transcription initiation factor TFIID subunit 7; 39S ribosomal protein L19, mitochondrial; lys-63-specific deubiquitinase BRCC36-like; ubiquitin carboxyl-terminal hydrolase 46; 39S ribosomal protein L3, mitochondrial; histone acetyltransferase KAT8; aurora kinase B; cell division control protein 6 homolog; phenylalanine–tRNA ligase alpha subunit; ribonuclease P/MRP protein subunit POP5; DNA topoisomerase 3-beta-1; uncharacterized LOC724152; 60S ribosomal protein L19; protein AF-9; structural maintenance of chromosomes protein 5; rRNA methyltransferase 3, mitochondrial; DNA mismatch repair protein Mlh1; OTU domain-containing protein 5-B; sprT-like domain-containing protein Spartan; iron-sulfur cluster assembly 2 homolog, mitochondrial-like; adenylyltransferase and sulfurtransferase MOCS3; cell cycle checkpoint protein RAD17; transducin beta-like protein 3; tRNA:m(4)X modification enzyme TRM13 homolog; 39S ribosomal protein L23, mitochondrial; BRISC and BRCA1-A complex member 1-like; methionine aminopeptidase 1D, mitochondrial; nuclear envelope phosphatase-regulatory subunit 1; peptidoglycan-recognition protein 1; 39S ribosomal protein L21, mitochondrial; homeobox protein Nkx-6.1-like; DNA polymerase delta small subunit; protein prenyltransferase alpha subunit repeat-containing protein 1; probable DNA-directed RNA polymerases I and III subunit RPAC2; DNA-directed RNA polymerase I subunit RPA2; uncharacterized LOC725813; 39S ribosomal protein L37, mitochondrial; cell division cycle-associated protein 7-like; pre-mRNA-splicing factor 18; H/ACA ribonucleoprotein complex non-core subunit NAF1; polycomb protein Scm; FACT complex subunit Ssrp1; gem-associated protein 8-like; male-specific lethal 3 homolog; probable 39S ribosomal protein L49, mitochondrial; hairy/enhancer-of-split related with YRPW motif protein 1; putative lipoyltransferase 2, mitochondrial; homologous-pairing protein 2 homolog; DNA polymerase eta; ruvB-like 2; structural maintenance of chromosomes protein 6; E3 ubiquitin-protein ligase synoviolin A; probable elongator complex protein 3; ankyrin repeat and LEM domain-containing protein 2; survival motor neuron protein; uncharacterized LOC100578201; probable deoxyhypusine synthase; histone-lysine N-methyltransferase SETD2; ribonucleases P/MRP protein subunit POP1; protein CASC3 |
| Module 1 | GO: Cellular component | GO:0043229 | intracellular organelle | 215/474 | 447/1126 | 1.142591 | 0.0005873 | 0.0057046 | 0.0118626 | 406077 406084 408352 408493 408511 408548 408606 408611 408615 408632 408675 408711 408808 408837 408846 409049 409098 409119 409224 409313 409331 409340 409423 409479 409544 409586 409637 409723 409735 409765 409810 409867 409887 409906 409994 410027 410195 410200 410203 410217 410298 410343 410348 410387 410390 410395 410410 410459 410571 410757 410855 410856 410864 410907 410960 411044 411072 411103 411265 411279 411304 411327 411351 411433 411465 411603 411619 411640 411654 411799 411816 411833 411854 411865 411918 411985 412016 412072 412076 412077 412199 412267 412268 412278 412350 412377 412381 412409 412506 412589 412710 412750 412796 412827 412984 413055 413078 413181 413271 413340 413351 413404 413490 413499 413523 413548 413558 413650 413666 413688 413793 413878 413940 413963 550716 550895 550924 551090 551131 551158 551240 551383 551398 551427 551438 551470 551620 551642 551692 551745 551781 551784 551811 551825 551843 551961 551974 551997 552052 552167 552247 552253 552312 552353 552440 552449 552450 552484 552495 552522 552533 552563 552593 552601 552676 552696 552703 552733 552763 552780 552794 552815 685996 724152 724186 724205 724244 724315 724365 724559 724665 724708 724720 724855 724955 725012 725013 725023 725061 725144 725201 725220 725254 725515 725565 725719 725854 725905 726002 726007 726058 726137 726151 726204 726239 726422 726617 726731 726816 727192 727263 727284 100576104 100576131 100576600 100576667 100576784 100577491 100577816 100578040 100578252 100578450 100578704 100578879 100579040 | homeotic protein antennapedia; ecdysone receptor; elongation factor Ts, mitochondrial; uncharacterized LOC408493; protein preli-like; MICOS complex subunit Mic60; PRP3 pre-mRNA processing factor 3 homolog; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; spliceosome-associated protein CWC15 homolog; probable 39S ribosomal protein L24, mitochondrial; zinc finger matrin-type protein 5-like; prosaposin; cytochrome c oxidase subunit 5A, mitochondrial; tyrosine-protein phosphatase non-receptor type 14; paired amphipathic helix protein Sin3a; histone-lysine N-methyltransferase eggless; REST corepressor 3; ubiquinone biosynthesis protein COQ7; actin-related protein 2; YEATS domain-containing protein 2; UV excision repair protein RAD23 homolog B; male-specific lethal 1 homolog; 60S ribosomal protein L6; ruvB-like 1; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9; 60S ribosomal protein L23a; 40S ribosomal protein S12, mitochondrial; transcription initiation factor TFIID subunit 6; actin-related protein 6; anaphase-promoting complex subunit 4; FERM domain-containing protein 8; transcription factor Dp-1; transcription initiation factor TFIID subunit 2; INO80 complex subunit E; survival of motor neuron-related-splicing factor 30; COP9 signalosome complex subunit 6; mRNA turnover protein 4 homolog; POU domain protein CF1A; structural maintenance of chromosomes protein 3; conserved oligomeric Golgi complex subunit 6; nuclear pore complex protein Nup93-like; katanin p80 WD40 repeat-containing subunit B1; conserved oligomeric Golgi complex subunit 3; protein suppressor of forked; chromobox protein homolog 1-like; ATP-dependent DNA helicase PIF1; cyclin-H; DNA topoisomerase 3-alpha; circadian locomoter output cycles protein kaput; retinoblastoma-binding protein 5 homolog; 28S ribosomal protein S29, mitochondrial; nuclear pore complex protein Nup50; U4/U6 small nuclear ribonucleoprotein Prp31; myosin-IB; inhibitor of growth protein 1; cysteine-rich protein 2-binding protein-like; putative 28S ribosomal protein S5, mitochondrial; CXXC-type zinc finger protein 1-like; actin-related protein 8; reticulon-4-interacting protein 1, mitochondrial-like; G1/S-specific cyclin-E1; elongation factor G, mitochondrial; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; digestive organ expansion factor homolog; double-strand-break repair protein rad21 homolog; DNA replication licensing factor Mcm2; probable tRNA N6-adenosine threonylcarbamoyltransferase; N-acetylgalactosaminyltransferase 7; cytoplasmic dynein 1 light intermediate chain 1; la protein homolog; heat shock factor protein; exportin-2; nuclear pore complex protein Nup205; histone-lysine N-methyltransferase SETD1; transforming growth factor beta regulator 1; DNA replication complex GINS protein PSF1-like; vacuolar protein-sorting-associated protein 36; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; transcriptional adapter 1-like; dynactin subunit 2; transcription initiation factor IIA subunit 1; tuberin; histone deacetylase 3; parafibromin; vacuolar protein-sorting-associated protein 25; mitochondrial import inner membrane translocase subunit TIM44; general vesicular transport factor p115; BRCA1-A complex subunit BRE-like; trafficking protein particle complex subunit 3; DNA topoisomerase 1; 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial; Bardet-Biedl syndrome 2 protein homolog; 28S ribosomal protein S30, mitochondrial; uncharacterized LOC413055; very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase; mortality factor 4-like protein 1; galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase I; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; clavesin-2-like; cell division cycle protein 23 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; photoreceptor-specific nuclear receptor; nuclear RNA export factor 1-like; peroxisome biogenesis factor 1; DNA-directed RNA polymerase III subunit RPC5; enhancer of polycomb homolog 1; glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial; importin-9; splicing factor 3A subunit 3; clathrin heavy chain; general transcription factor IIH subunit 1; cleavage stimulation factor subunit 2; conserved oligomeric Golgi complex subunit 8; protein max; 28S ribosomal protein S17, mitochondrial; uncharacterized LOC551240; protein MAK16 homolog A; DNA polymerase delta catalytic subunit; erlin-1-like; actin-related protein 1; general transcription factor IIF subunit 2; pre-mRNA-processing-splicing factor 8; GPI mannosyltransferase 4; protein FAM50 homolog; integrator complex subunit 10; pre-rRNA-processing protein TSR1 homolog; probable actin-related protein 2/3 complex subunit 2; graves disease carrier protein homolog; nuclear hormone receptor HR96; LETM1 and EF-hand domain-containing protein anon-60Da, mitochondrial; V-type proton ATPase subunit G; thioredoxin-like protein 4A; cleavage and polyadenylation specificity factor subunit 1; zinc finger protein 330 homolog; IWS1-like protein; nuclear factor NF-kappa-B p100 subunit; mitochondrial chaperone BCS1; RAD50-interacting protein 1; DNA-directed RNA polymerase III subunit RPC4; histone chaperone asf1; mediator of RNA polymerase II transcription subunit 6; DNA repair protein RAD51 homolog A; uncharacterized LOC552484; ATP-binding cassette sub-family D member 3; alpha-(1,6)-fucosyltransferase; GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase; transcription initiation factor TFIID subunit 7; 39S ribosomal protein L19, mitochondrial; lys-63-specific deubiquitinase BRCC36-like; 39S ribosomal protein L3, mitochondrial; histone acetyltransferase KAT8; KAT8 regulatory NSL complex subunit 2; aurora kinase B; cell division control protein 6 homolog; ribonuclease P/MRP protein subunit POP5; mediator of RNA polymerase II transcription subunit 14; high mobility group protein 20A-like; suppressor of variegation 3-9; uncharacterized LOC724152; 60S ribosomal protein L19; protein AF-9; structural maintenance of chromosomes protein 5; forkhead box protein D3-like; ragulator complex protein LAMTOR4 homolog; cell cycle checkpoint protein RAD17; protein SET; 39S ribosomal protein L23, mitochondrial; vacuolar protein sorting-associated protein 37B; BRISC and BRCA1-A complex member 1-like; KH domain-containing protein akap-1; zinc finger protein 25-like; sister chromatid cohesion protein DCC1; mediator of RNA polymerase II transcription subunit 26; uncharacterized LOC725061; uncharacterized LOC725144; 39S ribosomal protein L21, mitochondrial; homeobox protein Nkx-6.1-like; endothelial zinc finger protein induced by tumor necrosis factor alpha; zinc finger protein 543-like; nitric oxide synthase-interacting protein homolog; DNA-directed RNA polymerase I subunit RPA2; 39S ribosomal protein L37, mitochondrial; pre-mRNA-splicing factor 18; tyrosine-protein kinase hopscotch; polycomb protein Scm; FACT complex subunit Ssrp1; male-specific lethal 3 homolog; probable 39S ribosomal protein L49, mitochondrial; hairy/enhancer-of-split related with YRPW motif protein 1; putative lipoyltransferase 2, mitochondrial; vacuolar protein sorting-associated protein 37A; 39S ribosomal protein L48, mitochondrial; probable 28S ribosomal protein S26, mitochondrial; ruvB-like 2; structural maintenance of chromosomes protein 6; chromobox protein homolog 5; transcription factor Sox-10-like; INO80 complex subunit B; zinc finger protein 267-like; zinc finger protein 345-like; E3 ubiquitin-protein ligase synoviolin A; high mobility group B protein 6-like; survival motor neuron protein; gamma-tubulin complex component 6; rotatin; Ets at 97D ortholog; histone-lysine N-methyltransferase SETD2; tubulin delta chain-like; protein CASC3; chondroitin sulfate synthase 2 |
| Module 1 | GO: Cellular component | GO:1990234 | transferase complex | 31/474 | 47/1126 | 1.566837 | 0.0006520 | 0.0057046 | 0.0118626 | 408577 409279 409423 409544 409735 409810 409906 410459 410855 411072 411265 411985 412199 412219 412268 412377 413181 413351 413499 413793 551131 551470 551745 552353 552563 552696 552703 724244 726816 727192 100576667 | uncharacterized LOC408577; cullin-4A; male-specific lethal 1 homolog; ruvB-like 1; transcription initiation factor TFIID subunit 6; anaphase-promoting complex subunit 4; transcription initiation factor TFIID subunit 2; cyclin-H; retinoblastoma-binding protein 5 homolog; cysteine-rich protein 2-binding protein-like; CXXC-type zinc finger protein 1-like; histone-lysine N-methyltransferase SETD1; transcriptional adapter 1-like; ubiquitin conjugation factor E4 B; transcription initiation factor IIA subunit 1; parafibromin; mortality factor 4-like protein 1; integrator complex subunit 7; cell division cycle protein 23 homolog; enhancer of polycomb homolog 1; protein max; general transcription factor IIF subunit 2; integrator complex subunit 10; DNA-directed RNA polymerase III subunit RPC4; transcription initiation factor TFIID subunit 7; histone acetyltransferase KAT8; KAT8 regulatory NSL complex subunit 2; structural maintenance of chromosomes protein 5; ruvB-like 2; structural maintenance of chromosomes protein 6; E3 ubiquitin-protein ligase synoviolin A |
| Module 1 | GO: Biological process | GO:0006139 | nucleobase-containing compound metabolic process | 160/463 | 285/960 | 1.164033 | 0.0009101 | 0.0265200 | 0.0621362 | 406084 406112 408346 408363 408441 408474 408501 408527 408606 408611 408615 408632 408687 408815 408980 409049 409331 409340 409347 409392 409544 409586 409653 409696 409735 409839 409883 409887 409891 409915 410027 410076 410093 410203 410390 410395 410410 410459 410571 410749 410757 410806 410907 410941 411164 411167 411218 411279 411433 411465 411640 411649 411654 411719 411786 411833 412069 412072 412159 412200 412268 412350 412377 412410 412467 412750 413087 413139 413181 413340 413351 413404 413523 413548 413558 413688 413690 413718 413793 413794 413815 413821 413878 413963 414001 550673 550692 550885 550895 550924 551083 551093 551150 551240 551321 551398 551470 551497 551538 551540 551616 551620 551721 551745 551812 551825 551835 551855 551974 551986 552172 552303 552353 552449 552450 552458 552484 552529 552554 552563 552601 552696 552755 552763 552768 552780 552787 724205 724244 724247 724296 724424 724496 724559 724596 724635 724855 725220 725268 725330 725633 725719 725859 725905 725948 726007 726058 726086 726137 726204 726449 726583 726816 727192 100576866 100577491 100578201 100578450 100578852 100578879 | ecdysone receptor; period circadian protein; UPF0396 protein CG6066; adenylate cyclase 3; adenosine kinase 1; apyrase; serine/threonine-protein kinase/endoribonuclease IRE1; pre-mRNA-splicing factor SPF27; PRP3 pre-mRNA processing factor 3 homolog; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; spliceosome-associated protein CWC15 homolog; putative tRNA (cytidine(32)/guanosine(34)-2’-O)-methyltransferase; probable DNA mismatch repair protein Msh6; DNA oxidative demethylase ALKBH1; paired amphipathic helix protein Sin3a; YEATS domain-containing protein 2; UV excision repair protein RAD23 homolog B; U4/U6.U5 tri-snRNP-associated protein 1; ATPase WRNIP1-like; ruvB-like 1; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9; tRNA-splicing ligase RtcB homolog; splicing factor 45; transcription initiation factor TFIID subunit 6; regulator of nonsense transcripts 1; splicing factor 3A subunit 1; transcription factor Dp-1; cell division cycle and apoptosis regulator protein 1; RNA-binding protein 26; survival of motor neuron-related-splicing factor 30; probable uridine-cytidine kinase; geminin; POU domain protein CF1A; protein suppressor of forked; chromobox protein homolog 1-like; ATP-dependent DNA helicase PIF1; cyclin-H; DNA topoisomerase 3-alpha; squamous cell carcinoma antigen recognized by T-cells 3; circadian locomoter output cycles protein kaput; histidine–tRNA ligase, cytoplasmic; U4/U6 small nuclear ribonucleoprotein Prp31; protein MTO1 homolog, mitochondrial; DNA-directed RNA polymerase III subunit RPC3; cysteine–tRNA ligase, cytoplasmic; WW domain-binding protein 11; actin-related protein 8; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; DNA replication licensing factor Mcm2; alkylated DNA repair protein alkB homolog 8; probable tRNA N6-adenosine threonylcarbamoyltransferase; probable queuine tRNA-ribosyltransferase; general transcription factor IIE subunit 1; la protein homolog; UTP–glucose-1-phosphate uridylyltransferase; DNA replication complex GINS protein PSF1-like; pre-mRNA-splicing factor SYF1; threonine–tRNA ligase, cytoplasmic; transcription initiation factor IIA subunit 1; histone deacetylase 3; parafibromin; U3 small nucleolar RNA-associated protein 6 homolog; bifunctional purine biosynthesis protein PURH; DNA topoisomerase 1; tRNA (uracil-5-)-methyltransferase homolog A-like; probable phenylalanine–tRNA ligase, mitochondrial; mortality factor 4-like protein 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; photoreceptor-specific nuclear receptor; DNA-directed RNA polymerase III subunit RPC5; helicase SKI2W; probable glutamine–tRNA ligase; enhancer of polycomb homolog 1; translin; CCA tRNA nucleotidyltransferase 1, mitochondrial-like; histone deacetylase complex subunit SAP130-A-like; glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial; splicing factor 3A subunit 3; mRNA-capping enzyme; inosine-5’-monophosphate dehydrogenase 1b; TATA-box-binding protein; nicotinate phosphoribosyltransferase; general transcription factor IIH subunit 1; cleavage stimulation factor subunit 2; probable tRNA pseudouridine synthase 2; V-type proton ATPase catalytic subunit A; BRCA2-interacting transcriptional repressor EMSY; uncharacterized LOC551240; 39S ribosomal protein L44, mitochondrial; DNA polymerase delta catalytic subunit; general transcription factor IIF subunit 2; lariat debranching enzyme; lysine–tRNA ligase; replication factor C subunit 2; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1; pre-mRNA-processing-splicing factor 8; V-type proton ATPase subunit B; integrator complex subunit 10; mitochondrial tRNA-specific 2-thiouridylase 1; nuclear hormone receptor HR96; CTP synthase; exosome complex component rrp45-like; thioredoxin-like protein 4A; deoxyribose-phosphate aldolase; G patch domain-containing protein 1 homolog; RNA-binding protein cabeza-like; DNA-directed RNA polymerase III subunit RPC4; mediator of RNA polymerase II transcription subunit 6; DNA repair protein RAD51 homolog A; DNA-directed RNA polymerase II subunit RPB3; uncharacterized LOC552484; U6 snRNA-associated Sm-like protein LSm4; RNA 3’-terminal phosphate cyclase-like; transcription initiation factor TFIID subunit 7; lys-63-specific deubiquitinase BRCC36-like; histone acetyltransferase KAT8; probable GDP-L-fucose synthase; cell division control protein 6 homolog; phenylalanine–tRNA ligase alpha subunit; ribonuclease P/MRP protein subunit POP5; DNA topoisomerase 3-beta-1; protein AF-9; structural maintenance of chromosomes protein 5; rRNA methyltransferase 3, mitochondrial; DNA mismatch repair protein Mlh1; sprT-like domain-containing protein Spartan; adenylyltransferase and sulfurtransferase MOCS3; cell cycle checkpoint protein RAD17; transducin beta-like protein 3; tRNA:m(4)X modification enzyme TRM13 homolog; BRISC and BRCA1-A complex member 1-like; homeobox protein Nkx-6.1-like; S1 RNA-binding domain-containing protein 1; DNA polymerase delta small subunit; probable DNA-directed RNA polymerases I and III subunit RPAC2; DNA-directed RNA polymerase I subunit RPA2; cell division cycle-associated protein 7-like; pre-mRNA-splicing factor 18; H/ACA ribonucleoprotein complex non-core subunit NAF1; polycomb protein Scm; FACT complex subunit Ssrp1; gem-associated protein 8-like; male-specific lethal 3 homolog; hairy/enhancer-of-split related with YRPW motif protein 1; homologous-pairing protein 2 homolog; DNA polymerase eta; ruvB-like 2; structural maintenance of chromosomes protein 6; probable elongator complex protein 3; survival motor neuron protein; uncharacterized LOC100578201; histone-lysine N-methyltransferase SETD2; ribonucleases P/MRP protein subunit POP1; protein CASC3 |
| Module 1 | GO: Cellular component | GO:0005681 | spliceosomal complex | 8/474 | 8/1126 | 2.375527 | 0.0009527 | 0.0066688 | 0.0143521 | 408611 408632 413523 413548 413963 551620 551974 725905 | Sip1/TFIP11 interacting protein; spliceosome-associated protein CWC15 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; splicing factor 3A subunit 3; pre-mRNA-processing-splicing factor 8; thioredoxin-like protein 4A; pre-mRNA-splicing factor 18 |
| Module 1 | KEGG | KEGG:03460 | Fanconi anemia pathway | 11/497 | 12/1081 | 1.993796 | 0.0012792 | 0.0262233 | 0.0639592 | 409389 410571 411121 552450 552787 552851 724296 725418 725934 726583 726862 | ubiquitin specific protease-like; DNA topoisomerase 3-alpha; WD repeat-containing protein 48; DNA repair protein RAD51 homolog A; DNA topoisomerase 3-beta-1; crossover junction endonuclease EME1; DNA mismatch repair protein Mlh1; replication protein A 32 kDa subunit; replication protein A 70 kDa DNA-binding subunit; DNA polymerase eta; DNA repair protein REV1 |
| Module 1 | GO: Biological process | GO:0046483 | heterocycle metabolic process | 164/463 | 295/960 | 1.152689 | 0.0014783 | 0.0265200 | 0.0883095 | 406084 406112 408346 408363 408441 408474 408501 408527 408606 408611 408615 408632 408687 408815 408859 408980 409049 409331 409340 409347 409392 409544 409586 409653 409696 409735 409839 409883 409887 409891 409915 410027 410076 410093 410203 410390 410395 410410 410459 410571 410749 410757 410806 410907 410941 411164 411167 411218 411279 411433 411465 411640 411649 411654 411719 411786 411833 412069 412072 412159 412200 412268 412350 412377 412410 412467 412750 413087 413139 413181 413340 413351 413404 413523 413548 413558 413688 413690 413718 413793 413794 413815 413821 413878 413963 414001 494506 550673 550692 550885 550895 550924 551083 551093 551150 551240 551321 551398 551470 551497 551538 551540 551616 551620 551721 551745 551812 551825 551835 551855 551974 551986 552172 552303 552353 552449 552450 552458 552484 552529 552554 552563 552601 552663 552696 552755 552763 552768 552780 552787 724205 724244 724247 724296 724424 724496 724559 724596 724635 724855 725220 725268 725330 725633 725719 725859 725905 725948 726007 726058 726086 726137 726204 726449 726583 726754 726816 727192 100576866 100577491 100578201 100578450 100578852 100578879 | ecdysone receptor; period circadian protein; UPF0396 protein CG6066; adenylate cyclase 3; adenosine kinase 1; apyrase; serine/threonine-protein kinase/endoribonuclease IRE1; pre-mRNA-splicing factor SPF27; PRP3 pre-mRNA processing factor 3 homolog; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; spliceosome-associated protein CWC15 homolog; putative tRNA (cytidine(32)/guanosine(34)-2’-O)-methyltransferase; probable DNA mismatch repair protein Msh6; pyrroline-5-carboxylate reductase 2; DNA oxidative demethylase ALKBH1; paired amphipathic helix protein Sin3a; YEATS domain-containing protein 2; UV excision repair protein RAD23 homolog B; U4/U6.U5 tri-snRNP-associated protein 1; ATPase WRNIP1-like; ruvB-like 1; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9; tRNA-splicing ligase RtcB homolog; splicing factor 45; transcription initiation factor TFIID subunit 6; regulator of nonsense transcripts 1; splicing factor 3A subunit 1; transcription factor Dp-1; cell division cycle and apoptosis regulator protein 1; RNA-binding protein 26; survival of motor neuron-related-splicing factor 30; probable uridine-cytidine kinase; geminin; POU domain protein CF1A; protein suppressor of forked; chromobox protein homolog 1-like; ATP-dependent DNA helicase PIF1; cyclin-H; DNA topoisomerase 3-alpha; squamous cell carcinoma antigen recognized by T-cells 3; circadian locomoter output cycles protein kaput; histidine–tRNA ligase, cytoplasmic; U4/U6 small nuclear ribonucleoprotein Prp31; protein MTO1 homolog, mitochondrial; DNA-directed RNA polymerase III subunit RPC3; cysteine–tRNA ligase, cytoplasmic; WW domain-binding protein 11; actin-related protein 8; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; DNA replication licensing factor Mcm2; alkylated DNA repair protein alkB homolog 8; probable tRNA N6-adenosine threonylcarbamoyltransferase; probable queuine tRNA-ribosyltransferase; general transcription factor IIE subunit 1; la protein homolog; UTP–glucose-1-phosphate uridylyltransferase; DNA replication complex GINS protein PSF1-like; pre-mRNA-splicing factor SYF1; threonine–tRNA ligase, cytoplasmic; transcription initiation factor IIA subunit 1; histone deacetylase 3; parafibromin; U3 small nucleolar RNA-associated protein 6 homolog; bifunctional purine biosynthesis protein PURH; DNA topoisomerase 1; tRNA (uracil-5-)-methyltransferase homolog A-like; probable phenylalanine–tRNA ligase, mitochondrial; mortality factor 4-like protein 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; photoreceptor-specific nuclear receptor; DNA-directed RNA polymerase III subunit RPC5; helicase SKI2W; probable glutamine–tRNA ligase; enhancer of polycomb homolog 1; translin; CCA tRNA nucleotidyltransferase 1, mitochondrial-like; histone deacetylase complex subunit SAP130-A-like; glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial; splicing factor 3A subunit 3; mRNA-capping enzyme; heme oxygenase; inosine-5’-monophosphate dehydrogenase 1b; TATA-box-binding protein; nicotinate phosphoribosyltransferase; general transcription factor IIH subunit 1; cleavage stimulation factor subunit 2; probable tRNA pseudouridine synthase 2; V-type proton ATPase catalytic subunit A; BRCA2-interacting transcriptional repressor EMSY; uncharacterized LOC551240; 39S ribosomal protein L44, mitochondrial; DNA polymerase delta catalytic subunit; general transcription factor IIF subunit 2; lariat debranching enzyme; lysine–tRNA ligase; replication factor C subunit 2; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1; pre-mRNA-processing-splicing factor 8; V-type proton ATPase subunit B; integrator complex subunit 10; mitochondrial tRNA-specific 2-thiouridylase 1; nuclear hormone receptor HR96; CTP synthase; exosome complex component rrp45-like; thioredoxin-like protein 4A; deoxyribose-phosphate aldolase; G patch domain-containing protein 1 homolog; RNA-binding protein cabeza-like; DNA-directed RNA polymerase III subunit RPC4; mediator of RNA polymerase II transcription subunit 6; DNA repair protein RAD51 homolog A; DNA-directed RNA polymerase II subunit RPB3; uncharacterized LOC552484; U6 snRNA-associated Sm-like protein LSm4; RNA 3’-terminal phosphate cyclase-like; transcription initiation factor TFIID subunit 7; lys-63-specific deubiquitinase BRCC36-like; pyridoxal kinase; histone acetyltransferase KAT8; probable GDP-L-fucose synthase; cell division control protein 6 homolog; phenylalanine–tRNA ligase alpha subunit; ribonuclease P/MRP protein subunit POP5; DNA topoisomerase 3-beta-1; protein AF-9; structural maintenance of chromosomes protein 5; rRNA methyltransferase 3, mitochondrial; DNA mismatch repair protein Mlh1; sprT-like domain-containing protein Spartan; adenylyltransferase and sulfurtransferase MOCS3; cell cycle checkpoint protein RAD17; transducin beta-like protein 3; tRNA:m(4)X modification enzyme TRM13 homolog; BRISC and BRCA1-A complex member 1-like; homeobox protein Nkx-6.1-like; S1 RNA-binding domain-containing protein 1; DNA polymerase delta small subunit; probable DNA-directed RNA polymerases I and III subunit RPAC2; DNA-directed RNA polymerase I subunit RPA2; cell division cycle-associated protein 7-like; pre-mRNA-splicing factor 18; H/ACA ribonucleoprotein complex non-core subunit NAF1; polycomb protein Scm; FACT complex subunit Ssrp1; gem-associated protein 8-like; male-specific lethal 3 homolog; hairy/enhancer-of-split related with YRPW motif protein 1; homologous-pairing protein 2 homolog; DNA polymerase eta; kynurenine formamidase; ruvB-like 2; structural maintenance of chromosomes protein 6; probable elongator complex protein 3; survival motor neuron protein; uncharacterized LOC100578201; histone-lysine N-methyltransferase SETD2; ribonucleases P/MRP protein subunit POP1; protein CASC3 |
| Module 1 | GO: Biological process | GO:1901360 | organic cyclic compound metabolic process | 166/463 | 300/960 | 1.147300 | 0.0018598 | 0.0265200 | 0.0987528 | 406084 406112 408346 408363 408441 408474 408501 408527 408606 408611 408615 408622 408632 408687 408815 408859 408980 409049 409331 409340 409347 409392 409544 409586 409653 409696 409735 409839 409883 409887 409891 409915 410027 410076 410093 410203 410390 410395 410410 410459 410571 410749 410757 410806 410907 410941 411164 411167 411218 411279 411433 411465 411640 411649 411654 411719 411786 411833 412069 412072 412159 412200 412268 412350 412377 412410 412467 412750 413087 413139 413181 413340 413351 413404 413523 413548 413558 413688 413690 413718 413793 413794 413815 413821 413878 413963 414001 494506 550673 550692 550885 550895 550924 551083 551093 551150 551240 551321 551398 551470 551497 551538 551540 551616 551620 551721 551745 551812 551825 551835 551855 551974 551986 552172 552303 552353 552449 552450 552458 552484 552529 552554 552563 552601 552663 552696 552755 552763 552768 552780 552787 724205 724244 724247 724296 724424 724496 724559 724596 724635 724855 725018 725220 725268 725330 725633 725719 725859 725905 725948 726007 726058 726086 726137 726204 726449 726583 726754 726816 727192 100576866 100577491 100578201 100578450 100578852 100578879 | ecdysone receptor; period circadian protein; UPF0396 protein CG6066; adenylate cyclase 3; adenosine kinase 1; apyrase; serine/threonine-protein kinase/endoribonuclease IRE1; pre-mRNA-splicing factor SPF27; PRP3 pre-mRNA processing factor 3 homolog; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; protein henna; spliceosome-associated protein CWC15 homolog; putative tRNA (cytidine(32)/guanosine(34)-2’-O)-methyltransferase; probable DNA mismatch repair protein Msh6; pyrroline-5-carboxylate reductase 2; DNA oxidative demethylase ALKBH1; paired amphipathic helix protein Sin3a; YEATS domain-containing protein 2; UV excision repair protein RAD23 homolog B; U4/U6.U5 tri-snRNP-associated protein 1; ATPase WRNIP1-like; ruvB-like 1; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9; tRNA-splicing ligase RtcB homolog; splicing factor 45; transcription initiation factor TFIID subunit 6; regulator of nonsense transcripts 1; splicing factor 3A subunit 1; transcription factor Dp-1; cell division cycle and apoptosis regulator protein 1; RNA-binding protein 26; survival of motor neuron-related-splicing factor 30; probable uridine-cytidine kinase; geminin; POU domain protein CF1A; protein suppressor of forked; chromobox protein homolog 1-like; ATP-dependent DNA helicase PIF1; cyclin-H; DNA topoisomerase 3-alpha; squamous cell carcinoma antigen recognized by T-cells 3; circadian locomoter output cycles protein kaput; histidine–tRNA ligase, cytoplasmic; U4/U6 small nuclear ribonucleoprotein Prp31; protein MTO1 homolog, mitochondrial; DNA-directed RNA polymerase III subunit RPC3; cysteine–tRNA ligase, cytoplasmic; WW domain-binding protein 11; actin-related protein 8; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; DNA replication licensing factor Mcm2; alkylated DNA repair protein alkB homolog 8; probable tRNA N6-adenosine threonylcarbamoyltransferase; probable queuine tRNA-ribosyltransferase; general transcription factor IIE subunit 1; la protein homolog; UTP–glucose-1-phosphate uridylyltransferase; DNA replication complex GINS protein PSF1-like; pre-mRNA-splicing factor SYF1; threonine–tRNA ligase, cytoplasmic; transcription initiation factor IIA subunit 1; histone deacetylase 3; parafibromin; U3 small nucleolar RNA-associated protein 6 homolog; bifunctional purine biosynthesis protein PURH; DNA topoisomerase 1; tRNA (uracil-5-)-methyltransferase homolog A-like; probable phenylalanine–tRNA ligase, mitochondrial; mortality factor 4-like protein 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; photoreceptor-specific nuclear receptor; DNA-directed RNA polymerase III subunit RPC5; helicase SKI2W; probable glutamine–tRNA ligase; enhancer of polycomb homolog 1; translin; CCA tRNA nucleotidyltransferase 1, mitochondrial-like; histone deacetylase complex subunit SAP130-A-like; glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial; splicing factor 3A subunit 3; mRNA-capping enzyme; heme oxygenase; inosine-5’-monophosphate dehydrogenase 1b; TATA-box-binding protein; nicotinate phosphoribosyltransferase; general transcription factor IIH subunit 1; cleavage stimulation factor subunit 2; probable tRNA pseudouridine synthase 2; V-type proton ATPase catalytic subunit A; BRCA2-interacting transcriptional repressor EMSY; uncharacterized LOC551240; 39S ribosomal protein L44, mitochondrial; DNA polymerase delta catalytic subunit; general transcription factor IIF subunit 2; lariat debranching enzyme; lysine–tRNA ligase; replication factor C subunit 2; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1; pre-mRNA-processing-splicing factor 8; V-type proton ATPase subunit B; integrator complex subunit 10; mitochondrial tRNA-specific 2-thiouridylase 1; nuclear hormone receptor HR96; CTP synthase; exosome complex component rrp45-like; thioredoxin-like protein 4A; deoxyribose-phosphate aldolase; G patch domain-containing protein 1 homolog; RNA-binding protein cabeza-like; DNA-directed RNA polymerase III subunit RPC4; mediator of RNA polymerase II transcription subunit 6; DNA repair protein RAD51 homolog A; DNA-directed RNA polymerase II subunit RPB3; uncharacterized LOC552484; U6 snRNA-associated Sm-like protein LSm4; RNA 3’-terminal phosphate cyclase-like; transcription initiation factor TFIID subunit 7; lys-63-specific deubiquitinase BRCC36-like; pyridoxal kinase; histone acetyltransferase KAT8; probable GDP-L-fucose synthase; cell division control protein 6 homolog; phenylalanine–tRNA ligase alpha subunit; ribonuclease P/MRP protein subunit POP5; DNA topoisomerase 3-beta-1; protein AF-9; structural maintenance of chromosomes protein 5; rRNA methyltransferase 3, mitochondrial; DNA mismatch repair protein Mlh1; sprT-like domain-containing protein Spartan; adenylyltransferase and sulfurtransferase MOCS3; cell cycle checkpoint protein RAD17; transducin beta-like protein 3; tRNA:m(4)X modification enzyme TRM13 homolog; BRISC and BRCA1-A complex member 1-like; phosphomevalonate kinase; homeobox protein Nkx-6.1-like; S1 RNA-binding domain-containing protein 1; DNA polymerase delta small subunit; probable DNA-directed RNA polymerases I and III subunit RPAC2; DNA-directed RNA polymerase I subunit RPA2; cell division cycle-associated protein 7-like; pre-mRNA-splicing factor 18; H/ACA ribonucleoprotein complex non-core subunit NAF1; polycomb protein Scm; FACT complex subunit Ssrp1; gem-associated protein 8-like; male-specific lethal 3 homolog; hairy/enhancer-of-split related with YRPW motif protein 1; homologous-pairing protein 2 homolog; DNA polymerase eta; kynurenine formamidase; ruvB-like 2; structural maintenance of chromosomes protein 6; probable elongator complex protein 3; survival motor neuron protein; uncharacterized LOC100578201; histone-lysine N-methyltransferase SETD2; ribonucleases P/MRP protein subunit POP1; protein CASC3 |
| Module 1 | GO: Biological process | GO:0006725 | cellular aromatic compound metabolic process | 164/463 | 297/960 | 1.144927 | 0.0023156 | 0.0265200 | 0.1038832 | 406084 406112 408346 408363 408441 408474 408501 408527 408606 408611 408615 408622 408632 408687 408815 408980 409049 409331 409340 409347 409392 409544 409586 409653 409696 409735 409839 409883 409887 409891 409915 410027 410076 410093 410203 410390 410395 410410 410459 410571 410749 410757 410806 410907 410941 411164 411167 411218 411279 411433 411465 411640 411649 411654 411719 411786 411833 412069 412072 412159 412200 412268 412350 412377 412410 412467 412750 413087 413139 413181 413340 413351 413404 413523 413548 413558 413688 413690 413718 413793 413794 413815 413821 413878 413963 414001 494506 550673 550692 550885 550895 550924 551083 551093 551150 551240 551321 551398 551470 551497 551538 551540 551616 551620 551721 551745 551812 551825 551835 551855 551974 551986 552172 552303 552353 552449 552450 552458 552484 552529 552554 552563 552601 552663 552696 552755 552763 552768 552780 552787 724205 724244 724247 724296 724424 724496 724559 724596 724635 724855 725220 725268 725330 725633 725719 725859 725905 725948 726007 726058 726086 726137 726204 726449 726583 726754 726816 727192 100576866 100577491 100578201 100578450 100578852 100578879 | ecdysone receptor; period circadian protein; UPF0396 protein CG6066; adenylate cyclase 3; adenosine kinase 1; apyrase; serine/threonine-protein kinase/endoribonuclease IRE1; pre-mRNA-splicing factor SPF27; PRP3 pre-mRNA processing factor 3 homolog; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; protein henna; spliceosome-associated protein CWC15 homolog; putative tRNA (cytidine(32)/guanosine(34)-2’-O)-methyltransferase; probable DNA mismatch repair protein Msh6; DNA oxidative demethylase ALKBH1; paired amphipathic helix protein Sin3a; YEATS domain-containing protein 2; UV excision repair protein RAD23 homolog B; U4/U6.U5 tri-snRNP-associated protein 1; ATPase WRNIP1-like; ruvB-like 1; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9; tRNA-splicing ligase RtcB homolog; splicing factor 45; transcription initiation factor TFIID subunit 6; regulator of nonsense transcripts 1; splicing factor 3A subunit 1; transcription factor Dp-1; cell division cycle and apoptosis regulator protein 1; RNA-binding protein 26; survival of motor neuron-related-splicing factor 30; probable uridine-cytidine kinase; geminin; POU domain protein CF1A; protein suppressor of forked; chromobox protein homolog 1-like; ATP-dependent DNA helicase PIF1; cyclin-H; DNA topoisomerase 3-alpha; squamous cell carcinoma antigen recognized by T-cells 3; circadian locomoter output cycles protein kaput; histidine–tRNA ligase, cytoplasmic; U4/U6 small nuclear ribonucleoprotein Prp31; protein MTO1 homolog, mitochondrial; DNA-directed RNA polymerase III subunit RPC3; cysteine–tRNA ligase, cytoplasmic; WW domain-binding protein 11; actin-related protein 8; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; DNA replication licensing factor Mcm2; alkylated DNA repair protein alkB homolog 8; probable tRNA N6-adenosine threonylcarbamoyltransferase; probable queuine tRNA-ribosyltransferase; general transcription factor IIE subunit 1; la protein homolog; UTP–glucose-1-phosphate uridylyltransferase; DNA replication complex GINS protein PSF1-like; pre-mRNA-splicing factor SYF1; threonine–tRNA ligase, cytoplasmic; transcription initiation factor IIA subunit 1; histone deacetylase 3; parafibromin; U3 small nucleolar RNA-associated protein 6 homolog; bifunctional purine biosynthesis protein PURH; DNA topoisomerase 1; tRNA (uracil-5-)-methyltransferase homolog A-like; probable phenylalanine–tRNA ligase, mitochondrial; mortality factor 4-like protein 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; photoreceptor-specific nuclear receptor; DNA-directed RNA polymerase III subunit RPC5; helicase SKI2W; probable glutamine–tRNA ligase; enhancer of polycomb homolog 1; translin; CCA tRNA nucleotidyltransferase 1, mitochondrial-like; histone deacetylase complex subunit SAP130-A-like; glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial; splicing factor 3A subunit 3; mRNA-capping enzyme; heme oxygenase; inosine-5’-monophosphate dehydrogenase 1b; TATA-box-binding protein; nicotinate phosphoribosyltransferase; general transcription factor IIH subunit 1; cleavage stimulation factor subunit 2; probable tRNA pseudouridine synthase 2; V-type proton ATPase catalytic subunit A; BRCA2-interacting transcriptional repressor EMSY; uncharacterized LOC551240; 39S ribosomal protein L44, mitochondrial; DNA polymerase delta catalytic subunit; general transcription factor IIF subunit 2; lariat debranching enzyme; lysine–tRNA ligase; replication factor C subunit 2; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1; pre-mRNA-processing-splicing factor 8; V-type proton ATPase subunit B; integrator complex subunit 10; mitochondrial tRNA-specific 2-thiouridylase 1; nuclear hormone receptor HR96; CTP synthase; exosome complex component rrp45-like; thioredoxin-like protein 4A; deoxyribose-phosphate aldolase; G patch domain-containing protein 1 homolog; RNA-binding protein cabeza-like; DNA-directed RNA polymerase III subunit RPC4; mediator of RNA polymerase II transcription subunit 6; DNA repair protein RAD51 homolog A; DNA-directed RNA polymerase II subunit RPB3; uncharacterized LOC552484; U6 snRNA-associated Sm-like protein LSm4; RNA 3’-terminal phosphate cyclase-like; transcription initiation factor TFIID subunit 7; lys-63-specific deubiquitinase BRCC36-like; pyridoxal kinase; histone acetyltransferase KAT8; probable GDP-L-fucose synthase; cell division control protein 6 homolog; phenylalanine–tRNA ligase alpha subunit; ribonuclease P/MRP protein subunit POP5; DNA topoisomerase 3-beta-1; protein AF-9; structural maintenance of chromosomes protein 5; rRNA methyltransferase 3, mitochondrial; DNA mismatch repair protein Mlh1; sprT-like domain-containing protein Spartan; adenylyltransferase and sulfurtransferase MOCS3; cell cycle checkpoint protein RAD17; transducin beta-like protein 3; tRNA:m(4)X modification enzyme TRM13 homolog; BRISC and BRCA1-A complex member 1-like; homeobox protein Nkx-6.1-like; S1 RNA-binding domain-containing protein 1; DNA polymerase delta small subunit; probable DNA-directed RNA polymerases I and III subunit RPAC2; DNA-directed RNA polymerase I subunit RPA2; cell division cycle-associated protein 7-like; pre-mRNA-splicing factor 18; H/ACA ribonucleoprotein complex non-core subunit NAF1; polycomb protein Scm; FACT complex subunit Ssrp1; gem-associated protein 8-like; male-specific lethal 3 homolog; hairy/enhancer-of-split related with YRPW motif protein 1; homologous-pairing protein 2 homolog; DNA polymerase eta; kynurenine formamidase; ruvB-like 2; structural maintenance of chromosomes protein 6; probable elongator complex protein 3; survival motor neuron protein; uncharacterized LOC100578201; histone-lysine N-methyltransferase SETD2; ribonucleases P/MRP protein subunit POP1; protein CASC3 |
| Module 1 | GO: Biological process | GO:0033554 | cellular response to stress | 23/463 | 31/960 | 1.538354 | 0.0026085 | 0.0265200 | 0.1038832 | 408815 408980 409340 409544 409696 410410 411279 550895 551427 551616 552450 552601 724152 724244 724296 724424 724559 724855 726058 726583 726816 727192 100576667 | probable DNA mismatch repair protein Msh6; DNA oxidative demethylase ALKBH1; UV excision repair protein RAD23 homolog B; ruvB-like 1; splicing factor 45; ATP-dependent DNA helicase PIF1; actin-related protein 8; general transcription factor IIH subunit 1; erlin-1-like; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1; DNA repair protein RAD51 homolog A; lys-63-specific deubiquitinase BRCC36-like; uncharacterized LOC724152; structural maintenance of chromosomes protein 5; DNA mismatch repair protein Mlh1; sprT-like domain-containing protein Spartan; cell cycle checkpoint protein RAD17; BRISC and BRCA1-A complex member 1-like; FACT complex subunit Ssrp1; DNA polymerase eta; ruvB-like 2; structural maintenance of chromosomes protein 6; E3 ubiquitin-protein ligase synoviolin A |
| Module 1 | GO: Cellular component | GO:0043233 | organelle lumen | 45/474 | 78/1126 | 1.370497 | 0.0029394 | 0.0146972 | 0.0236060 | 408511 409049 409423 409544 409735 409906 409994 410200 410395 410459 410855 410856 411072 411265 411279 411433 411465 411619 411985 412072 412077 412199 412268 412377 413181 413340 413351 413404 413793 550924 551131 551383 551470 551745 552353 552449 552563 552696 552703 552780 552794 726617 726731 726816 100576104 | protein preli-like; paired amphipathic helix protein Sin3a; male-specific lethal 1 homolog; ruvB-like 1; transcription initiation factor TFIID subunit 6; transcription initiation factor TFIID subunit 2; INO80 complex subunit E; mRNA turnover protein 4 homolog; chromobox protein homolog 1-like; cyclin-H; retinoblastoma-binding protein 5 homolog; 28S ribosomal protein S29, mitochondrial; cysteine-rich protein 2-binding protein-like; CXXC-type zinc finger protein 1-like; actin-related protein 8; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; double-strand-break repair protein rad21 homolog; histone-lysine N-methyltransferase SETD1; DNA replication complex GINS protein PSF1-like; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; transcriptional adapter 1-like; transcription initiation factor IIA subunit 1; parafibromin; mortality factor 4-like protein 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; enhancer of polycomb homolog 1; cleavage stimulation factor subunit 2; protein max; protein MAK16 homolog A; general transcription factor IIF subunit 2; integrator complex subunit 10; DNA-directed RNA polymerase III subunit RPC4; mediator of RNA polymerase II transcription subunit 6; transcription initiation factor TFIID subunit 7; histone acetyltransferase KAT8; KAT8 regulatory NSL complex subunit 2; ribonuclease P/MRP protein subunit POP5; mediator of RNA polymerase II transcription subunit 14; 39S ribosomal protein L48, mitochondrial; probable 28S ribosomal protein S26, mitochondrial; ruvB-like 2; INO80 complex subunit B |
| Module 1 | GO: Cellular component | GO:0070013 | intracellular organelle lumen | 45/474 | 78/1126 | 1.370497 | 0.0029394 | 0.0146972 | 0.0236060 | 408511 409049 409423 409544 409735 409906 409994 410200 410395 410459 410855 410856 411072 411265 411279 411433 411465 411619 411985 412072 412077 412199 412268 412377 413181 413340 413351 413404 413793 550924 551131 551383 551470 551745 552353 552449 552563 552696 552703 552780 552794 726617 726731 726816 100576104 | protein preli-like; paired amphipathic helix protein Sin3a; male-specific lethal 1 homolog; ruvB-like 1; transcription initiation factor TFIID subunit 6; transcription initiation factor TFIID subunit 2; INO80 complex subunit E; mRNA turnover protein 4 homolog; chromobox protein homolog 1-like; cyclin-H; retinoblastoma-binding protein 5 homolog; 28S ribosomal protein S29, mitochondrial; cysteine-rich protein 2-binding protein-like; CXXC-type zinc finger protein 1-like; actin-related protein 8; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; double-strand-break repair protein rad21 homolog; histone-lysine N-methyltransferase SETD1; DNA replication complex GINS protein PSF1-like; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; transcriptional adapter 1-like; transcription initiation factor IIA subunit 1; parafibromin; mortality factor 4-like protein 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; enhancer of polycomb homolog 1; cleavage stimulation factor subunit 2; protein max; protein MAK16 homolog A; general transcription factor IIF subunit 2; integrator complex subunit 10; DNA-directed RNA polymerase III subunit RPC4; mediator of RNA polymerase II transcription subunit 6; transcription initiation factor TFIID subunit 7; histone acetyltransferase KAT8; KAT8 regulatory NSL complex subunit 2; ribonuclease P/MRP protein subunit POP5; mediator of RNA polymerase II transcription subunit 14; 39S ribosomal protein L48, mitochondrial; probable 28S ribosomal protein S26, mitochondrial; ruvB-like 2; INO80 complex subunit B |
| Module 1 | KEGG | KEGG:03420 | Nucleotide excision repair | 11/497 | 13/1081 | 1.840427 | 0.0048673 | 0.0665198 | 0.1622435 | 409279 409340 410459 412240 412593 550895 551398 551540 725330 725418 725934 | cullin-4A; UV excision repair protein RAD23 homolog B; cyclin-H; DNA repair protein complementing XP-A cells homolog; DNA damage-binding protein 1; general transcription factor IIH subunit 1; DNA polymerase delta catalytic subunit; replication factor C subunit 2; DNA polymerase delta small subunit; replication protein A 32 kDa subunit; replication protein A 70 kDa DNA-binding subunit |
| Module 1 | GO: Biological process | GO:0006325 | chromatin organization | 17/463 | 22/960 | 1.602199 | 0.0049039 | 0.0427343 | 0.1662997 | 409423 409544 409765 409994 411044 411279 411985 412077 412377 412746 413181 552440 552733 724665 726137 726816 100576104 | male-specific lethal 1 homolog; ruvB-like 1; actin-related protein 6; INO80 complex subunit E; inhibitor of growth protein 1; actin-related protein 8; histone-lysine N-methyltransferase SETD1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; parafibromin; calcineurin-binding protein cabin-1-like; mortality factor 4-like protein 1; histone chaperone asf1; aurora kinase B; protein SET; male-specific lethal 3 homolog; ruvB-like 2; INO80 complex subunit B |
| Module 1 | GO: Molecular function | GO:0101005 | ubiquitinyl hydrolase activity | 12/626 | 15/1400 | 1.789137 | 0.0057184 | 0.0729528 | 0.1369810 | 409387 409389 410162 411362 411572 411981 412644 413813 552324 552601 552660 724338 | ubiquitin carboxyl-terminal hydrolase; ubiquitin specific protease-like; ataxin-3-like; ubiquitin carboxyl-terminal hydrolase 8-like; ubiquitin carboxyl-terminal hydrolase 14; probable ubiquitin carboxyl-terminal hydrolase FAF-X; josephin-2; ubiquitin carboxyl-terminal hydrolase isozyme L5; ubiquitin carboxyl-terminal hydrolase 5; lys-63-specific deubiquitinase BRCC36-like; ubiquitin carboxyl-terminal hydrolase 46; OTU domain-containing protein 5-B |
| Module 1 | GO: Molecular function | GO:0019899 | enzyme binding | 19/626 | 27/1400 | 1.573778 | 0.0059210 | 0.0729528 | 0.1369810 | 409255 409279 409370 411245 411865 412708 412817 413208 413677 413735 413738 413940 550698 551256 551427 551741 724332 100577386 100578506 | active breakpoint cluster region-related protein; cullin-4A; brefeldin A-inhibited guanine nucleotide-exchange protein 1; dedicator of cytokinesis protein 7; exportin-2; calcyclin-binding protein; importin-4-like; rab11 family-interacting protein 2; rho guanine nucleotide exchange factor 10; glutamate–cysteine ligase regulatory subunit; uncharacterized LOC413738; importin-9; cyclin-Y; protein CNPPD1; erlin-1-like; rac guanine nucleotide exchange factor JJ; rho guanine nucleotide exchange factor 3-like; ankyrin repeat and LEM domain-containing protein 2; Rho-associated, coiled-coil containing protein kinase 2 |
| Module 1 | GO: Molecular function | GO:0008276 | protein methyltransferase activity | 6/626 | 6/1400 | 2.236422 | 0.0078868 | 0.0729528 | 0.1369810 | 409098 409833 411985 552772 685996 100578450 | histone-lysine N-methyltransferase eggless; hemK methyltransferase family member 1; histone-lysine N-methyltransferase SETD1; protein-lysine N-methyltransferase N6AMT2; suppressor of variegation 3-9; histone-lysine N-methyltransferase SETD2 |
| Module 1 | KEGG | KEGG:03040 | Spliceosome | 29/497 | 45/1081 | 1.401699 | 0.0084708 | 0.0868256 | 0.2117698 | 408527 408606 408632 409347 409647 409696 409866 409883 410027 410434 410907 411218 411538 412064 412159 413523 413548 413963 550656 551331 551620 551974 552027 552198 552444 552527 552529 725905 727087 | pre-mRNA-splicing factor SPF27; PRP3 pre-mRNA processing factor 3 homolog; spliceosome-associated protein CWC15 homolog; U4/U6.U5 tri-snRNP-associated protein 1; THO complex subunit 1; splicing factor 45; eukaryotic initiation factor 4A-III; splicing factor 3A subunit 1; survival of motor neuron-related-splicing factor 30; splicing factor 3B subunit 4; U4/U6 small nuclear ribonucleoprotein Prp31; WW domain-binding protein 11; pre-mRNA-splicing factor RBM22; probable U2 small nuclear ribonucleoprotein A’; pre-mRNA-splicing factor SYF1; pre-mRNA-processing factor 17; intron-binding protein aquarius; splicing factor 3A subunit 3; splicing factor U2AF 50 kDa subunit; splicing factor 3B subunit 1; pre-mRNA-processing-splicing factor 8; thioredoxin-like protein 4A; heterogeneous nuclear ribonucleoprotein A1, A2/B1 homolog; pleiotropic regulator 1; polyglutamine-binding protein 1; cell division cycle 5-like protein; U6 snRNA-associated Sm-like protein LSm4; pre-mRNA-splicing factor 18; serine/arginine-rich splicing factor 7 |
| Module 1 | GO: Molecular function | GO:0005524 | ATP binding | 105/626 | 200/1400 | 1.174121 | 0.0104471 | 0.0773082 | 0.1674913 | 406092 408501 408512 408533 408708 408815 408866 409028 409125 409178 409191 409198 409276 409296 409313 409392 409394 409408 409447 409498 409544 409577 409653 409756 409839 409866 410076 410190 410217 410273 410289 410410 410575 410806 410958 410960 411003 411068 411080 411167 411279 411450 411492 411582 411640 411758 411858 412190 412200 412446 412608 412610 412741 412747 413052 413126 413139 413152 413252 413493 413666 413690 413718 413759 413878 550694 550968 551093 551343 551386 551438 551470 551538 551540 551608 551616 551659 551721 551773 551835 552206 552253 552450 552469 552495 552554 552567 552733 552763 552768 552806 724193 724296 724440 724496 725018 725441 725720 726002 726742 726816 727113 100577120 100577699 100578506 | cGMP-dependent protein kinase foraging; serine/threonine-protein kinase/endoribonuclease IRE1; origin recognition complex subunit 1; mitogen-activated protein kinase kinase kinase 15; ubiquitin-conjugating enzyme E2 S; probable DNA mismatch repair protein Msh6; uncharacterized aarF domain-containing protein kinase 1; probable ATP-dependent RNA helicase YTHDC2; mitogen-activated protein kinase kinase kinase 4; ATP-dependent zinc metalloprotease YME1 homolog; SUMO-activating enzyme subunit 2; 26S protease regulatory subunit 6A-B; phosphatidylinositol 5-phosphate 4-kinase type-2 alpha; T-complex protein 1 subunit gamma; actin-related protein 2; ATPase WRNIP1-like; NEDD8-activating enzyme E1 catalytic subunit; nuclear valosin-containing protein-like; DNA repair and recombination protein RAD54-like; kinase suppressor of Ras 2; ruvB-like 1; 5’-AMP-activated protein kinase catalytic subunit alpha-2; tRNA-splicing ligase RtcB homolog; vacuolar protein sorting-associated protein 4B; regulator of nonsense transcripts 1; eukaryotic initiation factor 4A-III; probable uridine-cytidine kinase; tyrosine-protein kinase Dnt; structural maintenance of chromosomes protein 3; transcription termination factor 2; paraplegin; ATP-dependent DNA helicase PIF1; dual specificity mitogen-activated protein kinase kinase 4; histidine–tRNA ligase, cytoplasmic; ubiquitin-like modifier-activating enzyme 1; myosin-IB; spermatogenesis-associated protein 5; kinesin 4A; serine/threonine-protein kinase greatwall; cysteine–tRNA ligase, cytoplasmic; actin-related protein 8; probable ATP-dependent RNA helicase DDX43; ATP-dependent RNA helicase DHX36; tyrosine-protein kinase Fer; DNA replication licensing factor Mcm2; N-terminal kinase-like protein; ATPase family AAA domain-containing protein 1-B; probable ATP-dependent RNA helicase DDX47; threonine–tRNA ligase, cytoplasmic; SCY1-like protein 2; serine/threonine-protein kinase D3; glutathione synthetase; manganese-transporting ATPase 13A1; receptor interacting protein kinase 5; uncharacterized LOC413052; serine/threonine-protein kinase TAO1; probable phenylalanine–tRNA ligase, mitochondrial; LIM domain kinase 1; ATP-binding cassette sub-family F member 2; inhibitor of nuclear factor kappa-B kinase subunit epsilon; peroxisome biogenesis factor 1; helicase SKI2W; probable glutamine–tRNA ligase; SCY1-like protein 2; glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial; T-complex protein 1 subunit eta; heat shock protein 75 kDa, mitochondrial; V-type proton ATPase catalytic subunit A; 26S protease regulatory subunit 7; 26S protease regulatory subunit 10B; actin-related protein 1; general transcription factor IIF subunit 2; lysine–tRNA ligase; replication factor C subunit 2; serine/threonine-protein kinase pelle; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1; serine/threonine-protein kinase STE20; V-type proton ATPase subunit B; serine/threonine-protein kinase VRK1-like; CTP synthase; putative ATP-dependent RNA helicase me31b; mitochondrial chaperone BCS1; DNA repair protein RAD51 homolog A; cyclin-dependent kinase 20-like; ATP-binding cassette sub-family D member 3; RNA 3’-terminal phosphate cyclase-like; DEAD-box helicase Dbp80; aurora kinase B; cell division control protein 6 homolog; phenylalanine–tRNA ligase alpha subunit; fidgetin-like protein 1; kinesin 8; DNA mismatch repair protein Mlh1; calcium-transporting ATPase type 2C member 1; adenylyltransferase and sulfurtransferase MOCS3; phosphomevalonate kinase; probable ATP-dependent RNA helicase DDX49; muscle, skeletal receptor tyrosine protein kinase-like; tyrosine-protein kinase hopscotch; chromosome transmission fidelity protein 18 homolog; ruvB-like 2; probable ATP-dependent RNA helicase spindle-E; dual specificity protein kinase TTK; 5-formyltetrahydrofolate cyclo-ligase; Rho-associated, coiled-coil containing protein kinase 2 |
| Module 1 | GO: Cellular component | GO:0005667 | transcription factor complex | 10/474 | 13/1126 | 1.827329 | 0.0114707 | 0.0457043 | 0.0622908 | 408615 409735 409887 409906 410459 410757 412268 551131 551470 552563 | nuclear transcription factor Y subunit gamma; transcription initiation factor TFIID subunit 6; transcription factor Dp-1; transcription initiation factor TFIID subunit 2; cyclin-H; circadian locomoter output cycles protein kaput; transcription initiation factor IIA subunit 1; protein max; general transcription factor IIF subunit 2; transcription initiation factor TFIID subunit 7 |
| Module 1 | GO: Biological process | GO:0022402 | cell cycle process | 12/463 | 15/960 | 1.658747 | 0.0119068 | 0.0907896 | 0.2860218 | 409810 411194 413499 552450 552733 724244 724819 725013 725144 726449 100577120 100577386 | anaphase-promoting complex subunit 4; MAU2 chromatid cohesion factor homolog; cell division cycle protein 23 homolog; DNA repair protein RAD51 homolog A; aurora kinase B; structural maintenance of chromosomes protein 5; mitotic spindle assembly checkpoint protein MAD1; sister chromatid cohesion protein DCC1; uncharacterized LOC725144; homologous-pairing protein 2 homolog; dual specificity protein kinase TTK; ankyrin repeat and LEM domain-containing protein 2 |
| Module 1 | GO: Cellular component | GO:0033202 | DNA helicase complex | 5/474 | 5/1126 | 2.375527 | 0.0130578 | 0.0457043 | 0.0622908 | 409544 409994 411279 726816 100576104 | ruvB-like 1; INO80 complex subunit E; actin-related protein 8; ruvB-like 2; INO80 complex subunit B |
| Module 1 | GO: Cellular component | GO:0044446 | intracellular organelle part | 110/474 | 225/1126 | 1.161369 | 0.0130584 | 0.0457043 | 0.0622908 | 408511 408548 408606 408611 408615 408632 408837 409049 409224 409313 409423 409544 409586 409723 409735 409810 409906 409994 410195 410200 410298 410343 410348 410387 410395 410459 410855 410856 410864 410907 410960 411072 411265 411279 411433 411465 411619 411799 411816 411918 411985 412072 412076 412077 412199 412267 412268 412377 412381 412409 412506 412589 413078 413181 413271 413340 413351 413404 413490 413499 413523 413548 413666 413793 413963 550716 550895 550924 551090 551131 551383 551438 551470 551620 551642 551745 551784 551811 551961 551974 552353 552449 552522 552533 552563 552601 552696 552703 552733 552780 552794 724186 724244 724365 724720 724855 725013 725905 726422 726617 726731 726816 727192 100576104 100576667 100577816 100578040 100578704 100578879 100579040 | protein preli-like; MICOS complex subunit Mic60; PRP3 pre-mRNA processing factor 3 homolog; Sip1/TFIP11 interacting protein; nuclear transcription factor Y subunit gamma; spliceosome-associated protein CWC15 homolog; cytochrome c oxidase subunit 5A, mitochondrial; paired amphipathic helix protein Sin3a; ubiquinone biosynthesis protein COQ7; actin-related protein 2; male-specific lethal 1 homolog; ruvB-like 1; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9; 40S ribosomal protein S12, mitochondrial; transcription initiation factor TFIID subunit 6; anaphase-promoting complex subunit 4; transcription initiation factor TFIID subunit 2; INO80 complex subunit E; COP9 signalosome complex subunit 6; mRNA turnover protein 4 homolog; conserved oligomeric Golgi complex subunit 6; nuclear pore complex protein Nup93-like; katanin p80 WD40 repeat-containing subunit B1; conserved oligomeric Golgi complex subunit 3; chromobox protein homolog 1-like; cyclin-H; retinoblastoma-binding protein 5 homolog; 28S ribosomal protein S29, mitochondrial; nuclear pore complex protein Nup50; U4/U6 small nuclear ribonucleoprotein Prp31; myosin-IB; cysteine-rich protein 2-binding protein-like; CXXC-type zinc finger protein 1-like; actin-related protein 8; ell-associated factor Eaf; probable cleavage and polyadenylation specificity factor subunit 2; double-strand-break repair protein rad21 homolog; N-acetylgalactosaminyltransferase 7; cytoplasmic dynein 1 light intermediate chain 1; nuclear pore complex protein Nup205; histone-lysine N-methyltransferase SETD1; DNA replication complex GINS protein PSF1-like; vacuolar protein-sorting-associated protein 36; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; transcriptional adapter 1-like; dynactin subunit 2; transcription initiation factor IIA subunit 1; parafibromin; vacuolar protein-sorting-associated protein 25; mitochondrial import inner membrane translocase subunit TIM44; general vesicular transport factor p115; BRCA1-A complex subunit BRE-like; very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase; mortality factor 4-like protein 1; galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase I; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial; integrator complex subunit 7; ribosome biogenesis protein WDR12 homolog; clavesin-2-like; cell division cycle protein 23 homolog; pre-mRNA-processing factor 17; intron-binding protein aquarius; peroxisome biogenesis factor 1; enhancer of polycomb homolog 1; splicing factor 3A subunit 3; clathrin heavy chain; general transcription factor IIH subunit 1; cleavage stimulation factor subunit 2; conserved oligomeric Golgi complex subunit 8; protein max; protein MAK16 homolog A; actin-related protein 1; general transcription factor IIF subunit 2; pre-mRNA-processing-splicing factor 8; GPI mannosyltransferase 4; integrator complex subunit 10; probable actin-related protein 2/3 complex subunit 2; graves disease carrier protein homolog; V-type proton ATPase subunit G; thioredoxin-like protein 4A; DNA-directed RNA polymerase III subunit RPC4; mediator of RNA polymerase II transcription subunit 6; alpha-(1,6)-fucosyltransferase; GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase; transcription initiation factor TFIID subunit 7; lys-63-specific deubiquitinase BRCC36-like; histone acetyltransferase KAT8; KAT8 regulatory NSL complex subunit 2; aurora kinase B; ribonuclease P/MRP protein subunit POP5; mediator of RNA polymerase II transcription subunit 14; 60S ribosomal protein L19; structural maintenance of chromosomes protein 5; ragulator complex protein LAMTOR4 homolog; vacuolar protein sorting-associated protein 37B; BRISC and BRCA1-A complex member 1-like; sister chromatid cohesion protein DCC1; pre-mRNA-splicing factor 18; vacuolar protein sorting-associated protein 37A; 39S ribosomal protein L48, mitochondrial; probable 28S ribosomal protein S26, mitochondrial; ruvB-like 2; structural maintenance of chromosomes protein 6; INO80 complex subunit B; E3 ubiquitin-protein ligase synoviolin A; gamma-tubulin complex component 6; rotatin; tubulin delta chain-like; protein CASC3; chondroitin sulfate synthase 2 |
| Module 1 | KEGG | KEGG:03430 | Mismatch repair | 7/497 | 8/1081 | 1.903169 | 0.0203963 | 0.1672496 | 0.4079260 | 408815 551398 551540 724296 725330 725418 725934 | probable DNA mismatch repair protein Msh6; DNA polymerase delta catalytic subunit; replication factor C subunit 2; DNA mismatch repair protein Mlh1; DNA polymerase delta small subunit; replication protein A 32 kDa subunit; replication protein A 70 kDa DNA-binding subunit |
| Module 1 | GO: Molecular function | GO:0060589 | nucleoside-triphosphatase regulator activity | 11/626 | 15/1400 | 1.640043 | 0.0235601 | 0.1452871 | 0.2134963 | 409230 409255 411909 412130 412278 550748 551546 724383 726432 727108 727370 | arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2; active breakpoint cluster region-related protein; ran GTPase-activating protein 1; ral GTPase-activating protein subunit beta; tuberin; stromal membrane-associated protein 1; BAG family molecular chaperone regulator 2; centaurin-gamma-1A; rab GTPase-binding effector protein 1; rho GTPase-activating protein 26; uncharacterized LOC727370 |
| Module 1 | GO: Molecular function | GO:0005543 | phospholipid binding | 12/626 | 17/1400 | 1.578651 | 0.0277456 | 0.1466554 | 0.2266136 | 408945 409154 410052 410297 411145 412076 413129 413460 413490 413831 414028 727370 | sorting nexin-27; sorting nexin-30-like; synaptotagmin 20; sorting nexin-16; nischarin; vacuolar protein-sorting-associated protein 36; sorting nexin-29; sorting nexin-13-like; clavesin-2-like; sorting nexin lst-4; sorting nexin-4-like; uncharacterized LOC727370 |
| Module 1 | GO: Cellular component | GO:0044427 | chromosomal part | 13/474 | 20/1126 | 1.544093 | 0.0318258 | 0.1012640 | 0.1307615 | 409049 409544 409994 410395 411279 412072 412077 413181 724244 725013 726816 727192 100576104 | paired amphipathic helix protein Sin3a; ruvB-like 1; INO80 complex subunit E; chromobox protein homolog 1-like; actin-related protein 8; DNA replication complex GINS protein PSF1-like; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; mortality factor 4-like protein 1; structural maintenance of chromosomes protein 5; sister chromatid cohesion protein DCC1; ruvB-like 2; structural maintenance of chromosomes protein 6; INO80 complex subunit B |
| Module 1 | GO: Molecular function | GO:0016772 | transferase activity, transferring phosphorus-containing groups | 56/626 | 104/1400 | 1.204227 | 0.0328882 | 0.1521079 | 0.2373289 | 406092 408441 408501 408533 408866 409125 409199 409255 409276 409498 409577 409860 410076 410190 410575 411080 411164 411582 411698 411758 411974 412069 412446 412608 412747 413052 413126 413152 413493 413688 413759 413815 413879 414001 551041 551240 551398 551500 551608 551659 551739 551773 552353 552458 552469 552663 552733 724496 725018 725330 725719 725720 726002 727071 100577120 100578506 | cGMP-dependent protein kinase foraging; adenosine kinase 1; serine/threonine-protein kinase/endoribonuclease IRE1; mitogen-activated protein kinase kinase kinase 15; uncharacterized aarF domain-containing protein kinase 1; mitogen-activated protein kinase kinase kinase 4; glycerol kinase; active breakpoint cluster region-related protein; phosphatidylinositol 5-phosphate 4-kinase type-2 alpha; kinase suppressor of Ras 2; 5’-AMP-activated protein kinase catalytic subunit alpha-2; GPI ethanolamine phosphate transferase 3; probable uridine-cytidine kinase; tyrosine-protein kinase Dnt; dual specificity mitogen-activated protein kinase kinase 4; serine/threonine-protein kinase greatwall; DNA-directed RNA polymerase III subunit RPC3; tyrosine-protein kinase Fer; ethanolaminephosphotransferase 1-like; N-terminal kinase-like protein; translation initiation factor eIF-2B subunit gamma; UTP–glucose-1-phosphate uridylyltransferase; SCY1-like protein 2; serine/threonine-protein kinase D3; receptor interacting protein kinase 5; uncharacterized LOC413052; serine/threonine-protein kinase TAO1; LIM domain kinase 1; inhibitor of nuclear factor kappa-B kinase subunit epsilon; DNA-directed RNA polymerase III subunit RPC5; SCY1-like protein 2; CCA tRNA nucleotidyltransferase 1, mitochondrial-like; CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase, mitochondrial; mRNA-capping enzyme; glycerol kinase; uncharacterized LOC551240; DNA polymerase delta catalytic subunit; phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform; serine/threonine-protein kinase pelle; serine/threonine-protein kinase STE20; DNA primase large subunit; serine/threonine-protein kinase VRK1-like; DNA-directed RNA polymerase III subunit RPC4; DNA-directed RNA polymerase II subunit RPB3; cyclin-dependent kinase 20-like; pyridoxal kinase; aurora kinase B; adenylyltransferase and sulfurtransferase MOCS3; phosphomevalonate kinase; DNA polymerase delta small subunit; DNA-directed RNA polymerase I subunit RPA2; muscle, skeletal receptor tyrosine protein kinase-like; tyrosine-protein kinase hopscotch; acylglycerol kinase, mitochondrial; dual specificity protein kinase TTK; Rho-associated, coiled-coil containing protein kinase 2 |
| Module 1 | GO: Biological process | GO:0006996 | organelle organization | 48/463 | 82/960 | 1.213718 | 0.0329815 | 0.2079691 | 0.5618279 | 409313 409423 409544 409755 409765 409810 409994 410200 410217 410348 410410 410571 411044 411194 411279 411472 411985 412077 412377 412409 412506 412746 412750 412827 413181 413196 413490 413499 413666 551784 552026 552253 552288 552440 552714 552733 552787 724244 724665 724819 725013 726137 726449 726816 100576104 100577386 100577816 100578040 | actin-related protein 2; male-specific lethal 1 homolog; ruvB-like 1; beta-parvin; actin-related protein 6; anaphase-promoting complex subunit 4; INO80 complex subunit E; mRNA turnover protein 4 homolog; structural maintenance of chromosomes protein 3; katanin p80 WD40 repeat-containing subunit B1; ATP-dependent DNA helicase PIF1; DNA topoisomerase 3-alpha; inhibitor of growth protein 1; MAU2 chromatid cohesion factor homolog; actin-related protein 8; dynamin-1-like protein; histone-lysine N-methyltransferase SETD1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; parafibromin; mitochondrial import inner membrane translocase subunit TIM44; general vesicular transport factor p115; calcineurin-binding protein cabin-1-like; DNA topoisomerase 1; Bardet-Biedl syndrome 2 protein homolog; mortality factor 4-like protein 1; ribosome maturation protein SBDS; clavesin-2-like; cell division cycle protein 23 homolog; peroxisome biogenesis factor 1; probable actin-related protein 2/3 complex subunit 2; F-actin-uncapping protein LRRC16A; mitochondrial chaperone BCS1; golgin-45; histone chaperone asf1; conserved oligomeric Golgi complex subunit 2; aurora kinase B; DNA topoisomerase 3-beta-1; structural maintenance of chromosomes protein 5; protein SET; mitotic spindle assembly checkpoint protein MAD1; sister chromatid cohesion protein DCC1; male-specific lethal 3 homolog; homologous-pairing protein 2 homolog; ruvB-like 2; INO80 complex subunit B; ankyrin repeat and LEM domain-containing protein 2; gamma-tubulin complex component 6; rotatin |
| Module 1 | GO: Biological process | GO:0000278 | mitotic cell cycle | 10/463 | 13/960 | 1.594949 | 0.0340933 | 0.2079691 | 0.5618279 | 409810 409887 411194 413499 552450 552733 724819 725013 100577120 100577386 | anaphase-promoting complex subunit 4; transcription factor Dp-1; MAU2 chromatid cohesion factor homolog; cell division cycle protein 23 homolog; DNA repair protein RAD51 homolog A; aurora kinase B; mitotic spindle assembly checkpoint protein MAD1; sister chromatid cohesion protein DCC1; dual specificity protein kinase TTK; ankyrin repeat and LEM domain-containing protein 2 |
| Module 1 | GO: Cellular component | GO:0044798 | nuclear transcription factor complex | 8/474 | 11/1126 | 1.727656 | 0.0396861 | 0.1157513 | 0.1579613 | 408615 409735 409906 410459 412268 551131 551470 552563 | nuclear transcription factor Y subunit gamma; transcription initiation factor TFIID subunit 6; transcription initiation factor TFIID subunit 2; cyclin-H; transcription initiation factor IIA subunit 1; protein max; general transcription factor IIF subunit 2; transcription initiation factor TFIID subunit 7 |
| Module 1 | GO: Molecular function | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 10/626 | 14/1400 | 1.597444 | 0.0399149 | 0.1640946 | 0.2595882 | 408924 408969 409532 412350 412467 412484 413631 413878 725158 726754 | peptidoglycan-recognition protein LC; aminoacylase-1-like; NAD-dependent protein deacetylase Sirt2; histone deacetylase 3; bifunctional purine biosynthesis protein PURH; peptidoglycan recognition protein S2; vanin-like protein 1; glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial; peptidoglycan-recognition protein 1; kynurenine formamidase |
| Module 1 | GO: Biological process | GO:1903047 | mitotic cell cycle process | 8/463 | 10/960 | 1.658747 | 0.0424858 | 0.2356032 | 0.6344923 | 409810 411194 413499 552450 724819 725013 100577120 100577386 | anaphase-promoting complex subunit 4; MAU2 chromatid cohesion factor homolog; cell division cycle protein 23 homolog; DNA repair protein RAD51 homolog A; mitotic spindle assembly checkpoint protein MAD1; sister chromatid cohesion protein DCC1; dual specificity protein kinase TTK; ankyrin repeat and LEM domain-containing protein 2 |
| Module 1 | GO: Molecular function | GO:0008047 | enzyme activator activity | 11/626 | 16/1400 | 1.537540 | 0.0453823 | 0.1679144 | 0.2627395 | 409230 409255 411909 412130 412278 550748 724383 725550 726432 727108 727370 | arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2; active breakpoint cluster region-related protein; ran GTPase-activating protein 1; ral GTPase-activating protein subunit beta; tuberin; stromal membrane-associated protein 1; centaurin-gamma-1A; proteasome activator complex subunit 4A-like; rab GTPase-binding effector protein 1; rho GTPase-activating protein 26; uncharacterized LOC727370 |
| Module 1 | GO: Cellular component | GO:0030880 | RNA polymerase complex | 10/474 | 15/1126 | 1.583685 | 0.0475700 | 0.1275787 | 0.1666615 | 409735 409906 410459 412268 412377 413351 551470 551745 552353 552563 | transcription initiation factor TFIID subunit 6; transcription initiation factor TFIID subunit 2; cyclin-H; transcription initiation factor IIA subunit 1; parafibromin; integrator complex subunit 7; general transcription factor IIF subunit 2; integrator complex subunit 10; DNA-directed RNA polymerase III subunit RPC4; transcription initiation factor TFIID subunit 7 |
| Module 1 | KEGG | KEGG:00310 | Lysine degradation | 10/497 | 14/1081 | 1.553607 | 0.0487202 | 0.3329215 | 0.8120037 | 409098 410254 411070 411140 411985 552130 725458 726218 100578450 100578909 | histone-lysine N-methyltransferase eggless; glutaryl-CoA dehydrogenase, mitochondrial; histone-lysine N-methyltransferase, H3 lysine-79 specific; putative aldehyde dehydrogenase family 7 member A1 homolog; histone-lysine N-methyltransferase SETD1; alpha-aminoadipic semialdehyde synthase, mitochondrial; hydroxylysine kinase; acetyl-CoA acetyltransferase, mitochondrial; histone-lysine N-methyltransferase SETD2; procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| Module 2 | KEGG | KEGG:03010 | Ribosome | 41/221 | 55/1081 | 3.646318 | 0.0000000 | 0.0000000 | 0.0000000 | 406120 406126 408526 408799 409294 409326 409552 410188 411380 413137 413296 413875 550651 550711 551107 551125 551330 551418 551644 551867 551870 552097 552106 552241 552266 552272 552318 552445 552517 552564 552632 552774 724142 724233 724868 725647 725884 726013 726439 726789 727128 | ribosomal protein LP1; ribosomal protein S8; 60S ribosomal protein L4; 40S ribosomal protein S24; 60S ribosomal protein L23; 40S ribosomal protein S2; 40S ribosomal protein S10-like; 60S ribosomal protein L8; 60S ribosomal protein L30; 60S ribosomal protein L15; 40S ribosomal protein S3a; 60S ribosomal protein L31; 40S ribosomal protein S4; 60S acidic ribosomal protein P0; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; 40S ribosomal protein S13; 60S ribosomal protein L36; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S12; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L3; 60S ribosomal protein L13; 40S ribosomal protein S7; 60S ribosomal protein L22; 60S ribosomal protein L28; 60S ribosomal protein L24; 40S ribosomal protein S17; 40S ribosomal protein S11; 40S ribosomal protein S6; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; 40S ribosomal protein S16; 40S ribosomal protein S26; 60S ribosomal protein L34 |
| Module 2 | GO: Cellular component | GO:0005840 | ribosome | 39/210 | 57/1126 | 3.668672 | 0.0000000 | 0.0000000 | 0.0000000 | 406120 406126 408526 408799 409294 409326 410024 410188 411380 413137 413296 413875 413884 550651 550711 551107 551125 551330 551418 551644 551867 551870 552097 552106 552241 552272 552318 552517 552564 552632 552774 724233 724868 725647 725884 726013 726439 726789 727128 | ribosomal protein LP1; ribosomal protein S8; 60S ribosomal protein L4; 40S ribosomal protein S24; 60S ribosomal protein L23; 40S ribosomal protein S2; 40S ribosomal protein S19-like; 60S ribosomal protein L8; 60S ribosomal protein L30; 60S ribosomal protein L15; 40S ribosomal protein S3a; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; 40S ribosomal protein S4; 60S acidic ribosomal protein P0; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; 40S ribosomal protein S13; 60S ribosomal protein L36; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; 40S ribosomal protein S7; 60S ribosomal protein L22; 60S ribosomal protein L28; 40S ribosomal protein S17; 40S ribosomal protein S11; 40S ribosomal protein S6; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; 40S ribosomal protein S16; 40S ribosomal protein S26; 60S ribosomal protein L34 |
| Module 2 | GO: Cellular component | GO:0005737 | cytoplasm | 95/210 | 309/1126 | 1.648482 | 0.0000000 | 0.0000000 | 0.0000000 | 406120 406126 406152 408284 408353 408418 408424 408526 408650 408782 408799 408901 408986 408987 409023 409079 409258 409294 409326 409397 409802 409809 409825 409842 409978 410024 410026 410030 410092 410188 410627 411147 411380 411520 411765 411778 412083 412289 412308 412511 412554 412842 412913 412975 413137 413145 413296 413762 413875 413884 413889 544670 550651 550667 550711 551107 551125 551275 551330 551418 551644 551841 551867 551870 551960 552001 552097 552106 552118 552241 552272 552318 552517 552531 552564 552632 552774 552776 724233 724516 724868 725057 725105 725583 725647 725789 725884 726013 726205 726301 726439 726789 727128 100577081 100578006 | ribosomal protein LP1; ribosomal protein S8; juvenile hormone epoxide hydrolase 1; mitochondrial import receptor subunit TOM40 homolog 1-like; proteasome subunit beta type-7; AP-2 complex subunit mu; COP9 signalosome complex subunit 8; 60S ribosomal protein L4; inositol oxygenase; tubulin beta-1; 40S ribosomal protein S24; actin-related protein 2/3 complex subunit 1A; proteasome subunit alpha type-3; sortilin-related receptor; methylthioribose-1-phosphate isomerase; eukaryotic translation initiation factor 4E type 3-A-like; eukaryotic translation initiation factor 3 subunit I; 60S ribosomal protein L23; 40S ribosomal protein S2; signal peptidase complex catalytic subunit SEC11A; proteasome subunit alpha type-2; T-complex protein 1 subunit beta; T-complex protein 1 subunit epsilon; eukaryotic translation initiation factor 3 subunit M; proteasome subunit beta type-2-like; 40S ribosomal protein S19-like; 26S proteasome regulatory complex subunit p48A; protein SEC13 homolog; dual oxidase maturation factor 1; 60S ribosomal protein L8; putative aminopeptidase W07G4.4; AP-2 complex subunit alpha; 60S ribosomal protein L30; proteasome subunit beta type-4; rab GDP dissociation inhibitor beta; RNA-binding protein 8A; coatomer subunit gamma; cytosolic Fe-S cluster assembly factor NUBP1 homolog; flap endonuclease 1; importin subunit alpha-3; transmembrane emp24 domain-containing protein eca; sorting nexin-8-like; Golgi SNAP receptor complex member 2; E3 ubiquitin-protein ligase parkin; 60S ribosomal protein L15; eukaryotic translation initiation factor 3 subunit F; 40S ribosomal protein S3a; complement component 1 Q subcomponent-binding protein, mitochondrial; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; cytoplasmic tRNA 2-thiolation protein 1; elongation factor 1-alpha F2; 40S ribosomal protein S4; succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial; 60S acidic ribosomal protein P0; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; T-complex protein 1 subunit delta; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; 40S ribosomal protein S13; 60S ribosomal protein L36; eukaryotic translation initiation factor 3 subunit K; UDP-galactose translocator; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; pyrimidodiazepine synthase; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; 10 kDa heat shock protein, mitochondrial; 40S ribosomal protein S7; 60S ribosomal protein L22; 60S ribosomal protein L28; signal recognition particle subunit SRP68; 40S ribosomal protein S17; 39S ribosomal protein L50, mitochondrial-like; 40S ribosomal protein S11; coatomer subunit beta; UDP-sugar transporter UST74c-like; bridging integrator 3-like; 40S ribosomal protein S6; ragulator complex protein LAMTOR1; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; inosine triphosphate pyrophosphatase; actin-related protein 2/3 complex subunit 4; 40S ribosomal protein S16; 40S ribosomal protein S26; 60S ribosomal protein L34; dihydroorotate dehydrogenase (quinone), mitochondrial; inorganic pyrophosphatase |
| Module 2 | GO: Biological process | GO:0019538 | protein metabolic process | 68/199 | 188/960 | 1.744895 | 0.0000000 | 0.0000011 | 0.0000015 | 406120 406126 408293 408353 408424 408526 408799 408986 409234 409258 409294 409326 409397 409560 409770 409802 409842 409978 410024 410026 410188 410385 410664 410763 411090 411380 411458 411520 411695 412557 413137 413145 413213 413296 413875 413884 413889 550651 550901 551029 551107 551125 551330 551418 551644 551841 551867 551870 551960 552097 552106 552241 552272 552318 552517 552564 552632 552774 724233 724868 725647 725789 725884 726013 726439 726782 726789 727128 | ribosomal protein LP1; ribosomal protein S8; exostosin-1; proteasome subunit beta type-7; COP9 signalosome complex subunit 8; 60S ribosomal protein L4; 40S ribosomal protein S24; proteasome subunit alpha type-3; S-phase kinase-associated protein 1; eukaryotic translation initiation factor 3 subunit I; 60S ribosomal protein L23; 40S ribosomal protein S2; signal peptidase complex catalytic subunit SEC11A; eukaryotic translation initiation factor 4B; probable tubulin polyglutamylase TTLL1; proteasome subunit alpha type-2; eukaryotic translation initiation factor 3 subunit M; proteasome subunit beta type-2-like; 40S ribosomal protein S19-like; 26S proteasome regulatory complex subunit p48A; 60S ribosomal protein L8; methionine aminopeptidase 1; protein arginine N-methyltransferase 5; deoxyhypusine hydroxylase; tubulin–tyrosine ligase-like protein 12; 60S ribosomal protein L30; histone-arginine methyltransferase CARMER; proteasome subunit beta type-4; proteasome subunit beta type-1; diphthine methyl ester synthase; 60S ribosomal protein L15; eukaryotic translation initiation factor 3 subunit F; tubulin polyglutamylase TTLL4-like; 40S ribosomal protein S3a; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; cytoplasmic tRNA 2-thiolation protein 1; 40S ribosomal protein S4; protein phosphatase methylesterase 1; inhibitor of growth protein 5; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; 40S ribosomal protein S13; 60S ribosomal protein L36; eukaryotic translation initiation factor 3 subunit K; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; 40S ribosomal protein S7; 60S ribosomal protein L22; 60S ribosomal protein L28; 40S ribosomal protein S17; 40S ribosomal protein S11; 40S ribosomal protein S6; ragulator complex protein LAMTOR1; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; 40S ribosomal protein S16; PEST proteolytic signal-containing nuclear protein-like; 40S ribosomal protein S26; 60S ribosomal protein L34 |
| Module 2 | GO: Cellular component | GO:0043232 | intracellular non-membrane-bounded organelle | 48/210 | 125/1126 | 2.058971 | 0.0000000 | 0.0000003 | 0.0000006 | 406120 406126 408526 408585 408782 408799 408901 409294 409326 410024 410188 411380 412308 413137 413296 413686 413875 413884 550651 550711 551107 551125 551330 551418 551644 551867 551870 552097 552106 552241 552272 552318 552517 552564 552582 552632 552774 724233 724432 724868 725647 725884 726013 726301 726439 726789 727128 727247 | ribosomal protein LP1; ribosomal protein S8; 60S ribosomal protein L4; dynactin subunit 6; tubulin beta-1; 40S ribosomal protein S24; actin-related protein 2/3 complex subunit 1A; 60S ribosomal protein L23; 40S ribosomal protein S2; 40S ribosomal protein S19-like; 60S ribosomal protein L8; 60S ribosomal protein L30; flap endonuclease 1; 60S ribosomal protein L15; 40S ribosomal protein S3a; H/ACA ribonucleoprotein complex subunit 1; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; 40S ribosomal protein S4; 60S acidic ribosomal protein P0; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; 40S ribosomal protein S13; 60S ribosomal protein L36; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; 40S ribosomal protein S7; brahma-associated protein of 60 kDa; 60S ribosomal protein L22; 60S ribosomal protein L28; 40S ribosomal protein S17; ribosomal RNA processing protein 36 homolog; 40S ribosomal protein S11; 40S ribosomal protein S6; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; actin-related protein 2/3 complex subunit 4; 40S ribosomal protein S16; 40S ribosomal protein S26; 60S ribosomal protein L34; non-structural maintenance of chromosomes element 1 homolog |
| Module 2 | GO: Cellular component | GO:0044444 | cytoplasmic part | 67/210 | 214/1126 | 1.678727 | 0.0000004 | 0.0000033 | 0.0000071 | 406120 406126 406152 408284 408418 408526 408799 408987 409258 409294 409326 409397 409842 410024 410030 410092 410188 411147 411380 412083 412308 412554 412842 412913 412975 413137 413145 413296 413762 413875 413884 550651 550667 550711 551107 551125 551330 551418 551644 551841 551867 551870 551960 552001 552097 552106 552241 552272 552318 552517 552564 552632 552774 552776 724233 724516 724868 725057 725105 725647 725789 725884 726013 726439 726789 727128 100577081 | ribosomal protein LP1; ribosomal protein S8; juvenile hormone epoxide hydrolase 1; mitochondrial import receptor subunit TOM40 homolog 1-like; AP-2 complex subunit mu; 60S ribosomal protein L4; 40S ribosomal protein S24; sortilin-related receptor; eukaryotic translation initiation factor 3 subunit I; 60S ribosomal protein L23; 40S ribosomal protein S2; signal peptidase complex catalytic subunit SEC11A; eukaryotic translation initiation factor 3 subunit M; 40S ribosomal protein S19-like; protein SEC13 homolog; dual oxidase maturation factor 1; 60S ribosomal protein L8; AP-2 complex subunit alpha; 60S ribosomal protein L30; coatomer subunit gamma; flap endonuclease 1; transmembrane emp24 domain-containing protein eca; sorting nexin-8-like; Golgi SNAP receptor complex member 2; E3 ubiquitin-protein ligase parkin; 60S ribosomal protein L15; eukaryotic translation initiation factor 3 subunit F; 40S ribosomal protein S3a; complement component 1 Q subcomponent-binding protein, mitochondrial; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; 40S ribosomal protein S4; succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial; 60S acidic ribosomal protein P0; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; 40S ribosomal protein S13; 60S ribosomal protein L36; eukaryotic translation initiation factor 3 subunit K; UDP-galactose translocator; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; 40S ribosomal protein S7; 60S ribosomal protein L22; 60S ribosomal protein L28; signal recognition particle subunit SRP68; 40S ribosomal protein S17; 39S ribosomal protein L50, mitochondrial-like; 40S ribosomal protein S11; coatomer subunit beta; UDP-sugar transporter UST74c-like; 40S ribosomal protein S6; ragulator complex protein LAMTOR1; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; 40S ribosomal protein S16; 40S ribosomal protein S26; 60S ribosomal protein L34; dihydroorotate dehydrogenase (quinone), mitochondrial |
| Module 2 | GO: Biological process | GO:1901564 | organonitrogen compound metabolic process | 84/199 | 271/960 | 1.495299 | 0.0000012 | 0.0000287 | 0.0000589 | 406120 406126 408293 408353 408368 408424 408526 408799 408986 409023 409234 409258 409294 409326 409397 409487 409560 409770 409802 409842 409978 410024 410026 410122 410188 410385 410664 410763 411090 411307 411380 411458 411520 411695 412015 412557 413137 413145 413213 413296 413481 413854 413875 413884 413889 550651 550767 550901 551029 551107 551125 551330 551418 551644 551785 551841 551867 551870 551948 551960 551966 551983 552097 552106 552241 552272 552318 552517 552556 552564 552632 552774 724233 724868 725647 725789 725884 726013 726439 726782 726789 727128 727293 100577081 | ribosomal protein LP1; ribosomal protein S8; exostosin-1; proteasome subunit beta type-7; adenosylhomocysteinase; COP9 signalosome complex subunit 8; 60S ribosomal protein L4; 40S ribosomal protein S24; proteasome subunit alpha type-3; methylthioribose-1-phosphate isomerase; S-phase kinase-associated protein 1; eukaryotic translation initiation factor 3 subunit I; 60S ribosomal protein L23; 40S ribosomal protein S2; signal peptidase complex catalytic subunit SEC11A; probable glutamine-dependent NAD(+) synthetase; eukaryotic translation initiation factor 4B; probable tubulin polyglutamylase TTLL1; proteasome subunit alpha type-2; eukaryotic translation initiation factor 3 subunit M; proteasome subunit beta type-2-like; 40S ribosomal protein S19-like; 26S proteasome regulatory complex subunit p48A; glyceraldehyde-3-phosphate dehydrogenase 2; 60S ribosomal protein L8; methionine aminopeptidase 1; protein arginine N-methyltransferase 5; deoxyhypusine hydroxylase; tubulin–tyrosine ligase-like protein 12; mitochondrial enolase superfamily member 1-like; 60S ribosomal protein L30; histone-arginine methyltransferase CARMER; proteasome subunit beta type-4; proteasome subunit beta type-1; 6-pyruvoyl tetrahydrobiopterin synthase; diphthine methyl ester synthase; 60S ribosomal protein L15; eukaryotic translation initiation factor 3 subunit F; tubulin polyglutamylase TTLL4-like; 40S ribosomal protein S3a; probable chitinase 3; xylosyltransferase oxt; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; cytoplasmic tRNA 2-thiolation protein 1; 40S ribosomal protein S4; ribose 5-phosphate isomerase A; protein phosphatase methylesterase 1; inhibitor of growth protein 5; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; 6-phosphogluconolactonase; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; 40S ribosomal protein S13; 60S ribosomal protein L36; uracil phosphoribosyltransferase homolog; eukaryotic translation initiation factor 3 subunit K; multifunctional protein ADE2; thymidylate synthase; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; arginase-1; 40S ribosomal protein S7; 60S ribosomal protein L22; 60S ribosomal protein L28; 40S ribosomal protein S17; 40S ribosomal protein S11; 40S ribosomal protein S6; ragulator complex protein LAMTOR1; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; 40S ribosomal protein S16; PEST proteolytic signal-containing nuclear protein-like; 40S ribosomal protein S26; 60S ribosomal protein L34; guanine deaminase; dihydroorotate dehydrogenase (quinone), mitochondrial |
| Module 2 | GO: Cellular component | GO:0044391 | ribosomal subunit | 9/210 | 14/1126 | 3.446939 | 0.0001965 | 0.0013363 | 0.0027409 | 409326 410188 413296 551330 551418 551644 552272 552318 726013 | 40S ribosomal protein S2; 60S ribosomal protein L8; 40S ribosomal protein S3a; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; 40S ribosomal protein S23; 60S ribosomal protein L27a; 40S ribosomal protein S20 |
| Module 2 | GO: Biological process | GO:0044260 | cellular macromolecule metabolic process | 86/199 | 314/960 | 1.321256 | 0.0003197 | 0.0049016 | 0.0113287 | 406120 406126 408293 408353 408424 408526 408728 408799 408986 409234 409258 409294 409326 409397 409472 409560 409770 409802 409822 409842 409978 409979 410024 410174 410188 410385 410494 410664 410753 410763 411090 411380 411458 411520 411695 412308 412388 412557 413137 413145 413213 413296 413520 413875 413884 413889 550651 550901 551029 551107 551125 551212 551330 551418 551644 551734 551841 551867 551870 551960 552097 552106 552241 552272 552318 552517 552564 552599 552632 552774 724233 724412 724868 725352 725647 725789 725884 726013 726184 726359 726439 726782 726789 727128 727247 100578801 | ribosomal protein LP1; ribosomal protein S8; exostosin-1; proteasome subunit beta type-7; COP9 signalosome complex subunit 8; 60S ribosomal protein L4; mediator of RNA polymerase II transcription subunit 18; 40S ribosomal protein S24; proteasome subunit alpha type-3; S-phase kinase-associated protein 1; eukaryotic translation initiation factor 3 subunit I; 60S ribosomal protein L23; 40S ribosomal protein S2; signal peptidase complex catalytic subunit SEC11A; protein Smaug homolog 1; eukaryotic translation initiation factor 4B; probable tubulin polyglutamylase TTLL1; proteasome subunit alpha type-2; enhancer of split m7 protein-like; eukaryotic translation initiation factor 3 subunit M; proteasome subunit beta type-2-like; DNA replication licensing factor Mcm7; 40S ribosomal protein S19-like; cyclin-T; 60S ribosomal protein L8; methionine aminopeptidase 1; DNA-directed RNA polymerases I, II, and III subunit RPABC2; protein arginine N-methyltransferase 5; TATA-box-binding protein-like; deoxyhypusine hydroxylase; tubulin–tyrosine ligase-like protein 12; 60S ribosomal protein L30; histone-arginine methyltransferase CARMER; proteasome subunit beta type-4; proteasome subunit beta type-1; flap endonuclease 1; cell differentiation protein RCD1 homolog; diphthine methyl ester synthase; 60S ribosomal protein L15; eukaryotic translation initiation factor 3 subunit F; tubulin polyglutamylase TTLL4-like; 40S ribosomal protein S3a; mediator of RNA polymerase II transcription subunit 4; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; cytoplasmic tRNA 2-thiolation protein 1; 40S ribosomal protein S4; protein phosphatase methylesterase 1; inhibitor of growth protein 5; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; mediator of RNA polymerase II transcription subunit 16; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; DNA excision repair protein haywire; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; 40S ribosomal protein S13; 60S ribosomal protein L36; eukaryotic translation initiation factor 3 subunit K; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; 40S ribosomal protein S7; DNA-directed RNA polymerase II subunit RPB7; 60S ribosomal protein L22; 60S ribosomal protein L28; 40S ribosomal protein S17; muscle segmentation homeobox; 40S ribosomal protein S11; U6 snRNA-associated Sm-like protein LSm7; 40S ribosomal protein S6; ragulator complex protein LAMTOR1; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; uncharacterized LOC726184; protein SMG8; 40S ribosomal protein S16; PEST proteolytic signal-containing nuclear protein-like; 40S ribosomal protein S26; 60S ribosomal protein L34; non-structural maintenance of chromosomes element 1 homolog; uncharacterized LOC100578801 |
| Module 2 | GO: Cellular component | GO:0005839 | proteasome core complex | 6/210 | 9/1126 | 3.574603 | 0.0020066 | 0.0113707 | 0.0223894 | 408353 408986 409802 409978 411520 411695 | proteasome subunit beta type-7; proteasome subunit alpha type-3; proteasome subunit alpha type-2; proteasome subunit beta type-2-like; proteasome subunit beta type-4; proteasome subunit beta type-1 |
| Module 2 | GO: Cellular component | GO:1905368 | peptidase complex | 8/210 | 17/1126 | 2.523249 | 0.0066764 | 0.0324282 | 0.0677223 | 408353 408702 408986 409397 409802 409978 411520 411695 | proteasome subunit beta type-7; SAGA-associated factor 29; proteasome subunit alpha type-3; signal peptidase complex catalytic subunit SEC11A; proteasome subunit alpha type-2; proteasome subunit beta type-2-like; proteasome subunit beta type-4; proteasome subunit beta type-1 |
| Module 2 | KEGG | KEGG:00590 | Arachidonic acid metabolism | 4/221 | 5/1081 | 3.913122 | 0.0071690 | 0.1173348 | 0.2530455 | 409614 494523 551072 552304 | group XIIA secretory phospholipase A2; glutathione peroxidase-like 1; carbonyl reductase [NADPH] 1-like; glutathione S-transferase S1 |
| Module 2 | GO: Cellular component | GO:0000502 | proteasome complex | 6/210 | 11/1126 | 2.924675 | 0.0078927 | 0.0335440 | 0.0677431 | 408353 408986 409802 409978 411520 411695 | proteasome subunit beta type-7; proteasome subunit alpha type-3; proteasome subunit alpha type-2; proteasome subunit beta type-2-like; proteasome subunit beta type-4; proteasome subunit beta type-1 |
| Module 2 | KEGG | KEGG:03050 | Proteasome | 11/221 | 26/1081 | 2.069440 | 0.0085855 | 0.1173348 | 0.2530455 | 408353 408397 408986 409802 409978 410026 410072 411520 411695 551411 552319 | proteasome subunit beta type-7; 26S proteasome non-ATPase regulatory subunit 11; proteasome subunit alpha type-3; proteasome subunit alpha type-2; proteasome subunit beta type-2-like; 26S proteasome regulatory complex subunit p48A; 26S proteasome non-ATPase regulatory subunit 14; proteasome subunit beta type-4; proteasome subunit beta type-1; proteasome maturation protein; 26S proteasome non-ATPase regulatory subunit 6 |
| Module 2 | GO: Biological process | GO:1901576 | organic substance biosynthetic process | 73/199 | 286/960 | 1.231332 | 0.0114452 | 0.1111003 | 0.2445490 | 406120 406126 408293 408526 408728 408799 409023 409258 409294 409326 409472 409487 409560 409822 409842 409979 410024 410122 410174 410188 410494 410753 411380 412015 412308 412557 413137 413145 413296 413520 413655 413854 413875 413884 550651 551107 551125 551212 551330 551418 551644 551734 551841 551867 551870 551960 551966 551983 552097 552106 552241 552272 552318 552517 552556 552564 552599 552632 552774 724233 724412 724868 725105 725647 725884 726013 726184 726205 726439 726789 727128 100577081 100578801 | ribosomal protein LP1; ribosomal protein S8; exostosin-1; 60S ribosomal protein L4; mediator of RNA polymerase II transcription subunit 18; 40S ribosomal protein S24; methylthioribose-1-phosphate isomerase; eukaryotic translation initiation factor 3 subunit I; 60S ribosomal protein L23; 40S ribosomal protein S2; protein Smaug homolog 1; probable glutamine-dependent NAD(+) synthetase; eukaryotic translation initiation factor 4B; enhancer of split m7 protein-like; eukaryotic translation initiation factor 3 subunit M; DNA replication licensing factor Mcm7; 40S ribosomal protein S19-like; glyceraldehyde-3-phosphate dehydrogenase 2; cyclin-T; 60S ribosomal protein L8; DNA-directed RNA polymerases I, II, and III subunit RPABC2; TATA-box-binding protein-like; 60S ribosomal protein L30; 6-pyruvoyl tetrahydrobiopterin synthase; flap endonuclease 1; diphthine methyl ester synthase; 60S ribosomal protein L15; eukaryotic translation initiation factor 3 subunit F; 40S ribosomal protein S3a; mediator of RNA polymerase II transcription subunit 4; 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial; xylosyltransferase oxt; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; 40S ribosomal protein S4; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; mediator of RNA polymerase II transcription subunit 16; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; DNA excision repair protein haywire; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; 40S ribosomal protein S13; 60S ribosomal protein L36; eukaryotic translation initiation factor 3 subunit K; multifunctional protein ADE2; thymidylate synthase; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; arginase-1; 40S ribosomal protein S7; DNA-directed RNA polymerase II subunit RPB7; 60S ribosomal protein L22; 60S ribosomal protein L28; 40S ribosomal protein S17; muscle segmentation homeobox; 40S ribosomal protein S11; UDP-sugar transporter UST74c-like; 40S ribosomal protein S6; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; uncharacterized LOC726184; inosine triphosphate pyrophosphatase; 40S ribosomal protein S16; 40S ribosomal protein S26; 60S ribosomal protein L34; dihydroorotate dehydrogenase (quinone), mitochondrial; uncharacterized LOC100578801 |
| Module 2 | GO: Biological process | GO:0044249 | cellular biosynthetic process | 72/199 | 282/960 | 1.231690 | 0.0120761 | 0.1111003 | 0.2445490 | 406120 406126 408293 408526 408728 408799 409023 409258 409294 409326 409472 409487 409560 409822 409842 409979 410024 410122 410174 410188 410494 410753 411380 412015 412308 412557 413137 413145 413296 413520 413655 413875 413884 550651 551107 551125 551212 551330 551418 551644 551734 551841 551867 551870 551960 551966 551983 552097 552106 552241 552272 552318 552517 552556 552564 552599 552632 552774 724233 724412 724868 725105 725647 725884 726013 726184 726205 726439 726789 727128 100577081 100578801 | ribosomal protein LP1; ribosomal protein S8; exostosin-1; 60S ribosomal protein L4; mediator of RNA polymerase II transcription subunit 18; 40S ribosomal protein S24; methylthioribose-1-phosphate isomerase; eukaryotic translation initiation factor 3 subunit I; 60S ribosomal protein L23; 40S ribosomal protein S2; protein Smaug homolog 1; probable glutamine-dependent NAD(+) synthetase; eukaryotic translation initiation factor 4B; enhancer of split m7 protein-like; eukaryotic translation initiation factor 3 subunit M; DNA replication licensing factor Mcm7; 40S ribosomal protein S19-like; glyceraldehyde-3-phosphate dehydrogenase 2; cyclin-T; 60S ribosomal protein L8; DNA-directed RNA polymerases I, II, and III subunit RPABC2; TATA-box-binding protein-like; 60S ribosomal protein L30; 6-pyruvoyl tetrahydrobiopterin synthase; flap endonuclease 1; diphthine methyl ester synthase; 60S ribosomal protein L15; eukaryotic translation initiation factor 3 subunit F; 40S ribosomal protein S3a; mediator of RNA polymerase II transcription subunit 4; 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; 40S ribosomal protein S4; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; mediator of RNA polymerase II transcription subunit 16; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; DNA excision repair protein haywire; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; 40S ribosomal protein S13; 60S ribosomal protein L36; eukaryotic translation initiation factor 3 subunit K; multifunctional protein ADE2; thymidylate synthase; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; arginase-1; 40S ribosomal protein S7; DNA-directed RNA polymerase II subunit RPB7; 60S ribosomal protein L22; 60S ribosomal protein L28; 40S ribosomal protein S17; muscle segmentation homeobox; 40S ribosomal protein S11; UDP-sugar transporter UST74c-like; 40S ribosomal protein S6; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; uncharacterized LOC726184; inosine triphosphate pyrophosphatase; 40S ribosomal protein S16; 40S ribosomal protein S26; 60S ribosomal protein L34; dihydroorotate dehydrogenase (quinone), mitochondrial; uncharacterized LOC100578801 |
| Module 2 | GO: Molecular function | GO:0051082 | unfolded protein binding | 6/263 | 12/1400 | 2.661597 | 0.0139451 | 0.1673416 | 0.3824950 | 408928 409809 409825 411071 411936 551275 | heat shock protein 90; T-complex protein 1 subunit beta; T-complex protein 1 subunit epsilon; dnaJ protein homolog 1; prefoldin subunit 5; T-complex protein 1 subunit delta |
| Module 2 | KEGG | KEGG:00051 | Fructose and mannose metabolism | 4/221 | 6/1081 | 3.260935 | 0.0180656 | 0.1851723 | 0.3993447 | 409329 411307 411696 551968 | mannose-1-phosphate guanyltransferase beta; mitochondrial enolase superfamily member 1-like; GDP-mannose 4,6 dehydratase; aldose reductase-like |
| Module 2 | GO: Cellular component | GO:0043229 | intracellular organelle | 96/210 | 447/1126 | 1.151550 | 0.0293678 | 0.1109449 | 0.2128523 | 406120 406126 406152 408284 408353 408424 408526 408585 408702 408728 408782 408799 408901 408986 408987 409023 409294 409326 409356 409397 409802 409822 409978 409979 410024 410030 410092 410188 410494 410505 410636 410989 411380 411385 411520 411778 412083 412308 412511 412554 412913 412975 413137 413209 413296 413520 413686 413762 413842 413875 413884 550651 550667 550711 551029 551107 551125 551212 551330 551418 551607 551644 551841 551867 551870 552001 552097 552106 552241 552272 552318 552517 552564 552582 552632 552774 724233 724412 724432 724516 724868 725057 725102 725105 725647 725789 725884 726013 726301 726439 726789 727128 727247 100577081 100578801 100578888 | ribosomal protein LP1; ribosomal protein S8; juvenile hormone epoxide hydrolase 1; mitochondrial import receptor subunit TOM40 homolog 1-like; proteasome subunit beta type-7; COP9 signalosome complex subunit 8; 60S ribosomal protein L4; dynactin subunit 6; SAGA-associated factor 29; mediator of RNA polymerase II transcription subunit 18; tubulin beta-1; 40S ribosomal protein S24; actin-related protein 2/3 complex subunit 1A; proteasome subunit alpha type-3; sortilin-related receptor; methylthioribose-1-phosphate isomerase; 60S ribosomal protein L23; 40S ribosomal protein S2; small ribonucleoprotein particle protein B; signal peptidase complex catalytic subunit SEC11A; proteasome subunit alpha type-2; enhancer of split m7 protein-like; proteasome subunit beta type-2-like; DNA replication licensing factor Mcm7; 40S ribosomal protein S19-like; protein SEC13 homolog; dual oxidase maturation factor 1; 60S ribosomal protein L8; DNA-directed RNA polymerases I, II, and III subunit RPABC2; E3 ubiquitin-protein ligase RING1; CDGSH iron-sulfur domain-containing protein 2 homolog; menin; 60S ribosomal protein L30; ester hydrolase C11orf54 homolog; proteasome subunit beta type-4; RNA-binding protein 8A; coatomer subunit gamma; flap endonuclease 1; importin subunit alpha-3; transmembrane emp24 domain-containing protein eca; Golgi SNAP receptor complex member 2; E3 ubiquitin-protein ligase parkin; 60S ribosomal protein L15; dnaJ homolog subfamily C member 2; 40S ribosomal protein S3a; mediator of RNA polymerase II transcription subunit 4; H/ACA ribonucleoprotein complex subunit 1; complement component 1 Q subcomponent-binding protein, mitochondrial; splicing factor U2af 38 kDa subunit; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; 40S ribosomal protein S4; succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial; 60S acidic ribosomal protein P0; inhibitor of growth protein 5; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; mediator of RNA polymerase II transcription subunit 16; 40S ribosomal protein S3; 60S ribosomal protein L13a; DNA-directed RNA polymerase II subunit RPB11; 40S ribosomal protein S15; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; 40S ribosomal protein S13; 60S ribosomal protein L36; UDP-galactose translocator; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; 40S ribosomal protein S7; brahma-associated protein of 60 kDa; 60S ribosomal protein L22; 60S ribosomal protein L28; 40S ribosomal protein S17; muscle segmentation homeobox; ribosomal RNA processing protein 36 homolog; 39S ribosomal protein L50, mitochondrial-like; 40S ribosomal protein S11; coatomer subunit beta; protein dpy-30 homolog; UDP-sugar transporter UST74c-like; 40S ribosomal protein S6; ragulator complex protein LAMTOR1; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; actin-related protein 2/3 complex subunit 4; 40S ribosomal protein S16; 40S ribosomal protein S26; 60S ribosomal protein L34; non-structural maintenance of chromosomes element 1 homolog; dihydroorotate dehydrogenase (quinone), mitochondrial; uncharacterized LOC100578801; zinc finger protein 62 homolog |
| Module 2 | KEGG | KEGG:04150 | mTOR signaling pathway | 12/221 | 34/1081 | 1.726377 | 0.0299287 | 0.2454151 | 0.5292650 | 409050 409393 409560 409725 410030 411295 551198 552381 552720 725647 725789 725855 | segment polarity protein dishevelled homolog DVL-3; serine/threonine-protein kinase mTOR; eukaryotic translation initiation factor 4B; target of rapamycin complex subunit lst8; protein SEC13 homolog; V-type proton ATPase subunit D 1; serine/threonine-protein kinase STK11; neutral and basic amino acid transport protein rBAT; V-type proton ATPase subunit E; 40S ribosomal protein S6; ragulator complex protein LAMTOR1; ragulator complex protein LAMTOR3-A |
| Module 2 | GO: Biological process | GO:0043170 | macromolecule metabolic process | 98/199 | 414/960 | 1.141942 | 0.0304780 | 0.2098314 | 0.5029268 | 406120 406126 408293 408353 408424 408526 408728 408799 408986 409169 409234 409258 409294 409326 409397 409472 409560 409770 409802 409822 409842 409978 409979 410024 410026 410174 410188 410385 410494 410664 410753 410763 411076 411090 411380 411458 411520 411695 411778 412308 412388 412427 412557 413137 413145 413213 413296 413481 413520 413686 413842 413854 413875 413884 413889 550651 550901 551029 551107 551125 551212 551330 551418 551644 551734 551841 551867 551870 551960 551992 552097 552106 552241 552272 552318 552517 552559 552564 552599 552632 552774 724233 724412 724432 724868 725352 725647 725789 725884 726013 726184 726359 726439 726782 726789 727128 727247 100578801 | ribosomal protein LP1; ribosomal protein S8; exostosin-1; proteasome subunit beta type-7; COP9 signalosome complex subunit 8; 60S ribosomal protein L4; mediator of RNA polymerase II transcription subunit 18; 40S ribosomal protein S24; proteasome subunit alpha type-3; mRNA export factor; S-phase kinase-associated protein 1; eukaryotic translation initiation factor 3 subunit I; 60S ribosomal protein L23; 40S ribosomal protein S2; signal peptidase complex catalytic subunit SEC11A; protein Smaug homolog 1; eukaryotic translation initiation factor 4B; probable tubulin polyglutamylase TTLL1; proteasome subunit alpha type-2; enhancer of split m7 protein-like; eukaryotic translation initiation factor 3 subunit M; proteasome subunit beta type-2-like; DNA replication licensing factor Mcm7; 40S ribosomal protein S19-like; 26S proteasome regulatory complex subunit p48A; cyclin-T; 60S ribosomal protein L8; methionine aminopeptidase 1; DNA-directed RNA polymerases I, II, and III subunit RPABC2; protein arginine N-methyltransferase 5; TATA-box-binding protein-like; deoxyhypusine hydroxylase; UPF0553 protein C9orf64 homolog; tubulin–tyrosine ligase-like protein 12; 60S ribosomal protein L30; histone-arginine methyltransferase CARMER; proteasome subunit beta type-4; proteasome subunit beta type-1; RNA-binding protein 8A; flap endonuclease 1; cell differentiation protein RCD1 homolog; aubergine; diphthine methyl ester synthase; 60S ribosomal protein L15; eukaryotic translation initiation factor 3 subunit F; tubulin polyglutamylase TTLL4-like; 40S ribosomal protein S3a; probable chitinase 3; mediator of RNA polymerase II transcription subunit 4; H/ACA ribonucleoprotein complex subunit 1; splicing factor U2af 38 kDa subunit; xylosyltransferase oxt; 60S ribosomal protein L31; ubiquitin-40S ribosomal protein S27a; cytoplasmic tRNA 2-thiolation protein 1; 40S ribosomal protein S4; protein phosphatase methylesterase 1; inhibitor of growth protein 5; 60S ribosomal protein L9; 40S ribosomal protein S15Aa; mediator of RNA polymerase II transcription subunit 16; 40S ribosomal protein S3; 60S ribosomal protein L13a; 40S ribosomal protein S15; DNA excision repair protein haywire; probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase; 40S ribosomal protein S13; 60S ribosomal protein L36; eukaryotic translation initiation factor 3 subunit K; rRNA 2’-O-methyltransferase fibrillarin; 39S ribosomal protein L14, mitochondrial; uncharacterized LOC552106; 60S ribosomal protein L5; 40S ribosomal protein S23; 60S ribosomal protein L27a; 60S ribosomal protein L13; probable small nuclear ribonucleoprotein Sm D1; 40S ribosomal protein S7; DNA-directed RNA polymerase II subunit RPB7; 60S ribosomal protein L22; 60S ribosomal protein L28; 40S ribosomal protein S17; muscle segmentation homeobox; ribosomal RNA processing protein 36 homolog; 40S ribosomal protein S11; U6 snRNA-associated Sm-like protein LSm7; 40S ribosomal protein S6; ragulator complex protein LAMTOR1; 60S acidic ribosomal protein P2; 40S ribosomal protein S20; uncharacterized LOC726184; protein SMG8; 40S ribosomal protein S16; PEST proteolytic signal-containing nuclear protein-like; 40S ribosomal protein S26; 60S ribosomal protein L34; non-structural maintenance of chromosomes element 1 homolog; uncharacterized LOC100578801 |
| Module 2 | GO: Biological process | GO:0044248 | cellular catabolic process | 20/199 | 65/960 | 1.484345 | 0.0319309 | 0.2098314 | 0.5029268 | 406152 408353 408368 408650 408834 408986 409234 409472 409802 409978 410122 411307 411520 411695 412308 412388 725352 726205 726359 727293 | juvenile hormone epoxide hydrolase 1; proteasome subunit beta type-7; adenosylhomocysteinase; inositol oxygenase; beclin-1-like protein; proteasome subunit alpha type-3; S-phase kinase-associated protein 1; protein Smaug homolog 1; proteasome subunit alpha type-2; proteasome subunit beta type-2-like; glyceraldehyde-3-phosphate dehydrogenase 2; mitochondrial enolase superfamily member 1-like; proteasome subunit beta type-4; proteasome subunit beta type-1; flap endonuclease 1; cell differentiation protein RCD1 homolog; U6 snRNA-associated Sm-like protein LSm7; inosine triphosphate pyrophosphatase; protein SMG8; guanine deaminase |
| Module 2 | GO: Cellular component | GO:0030117 | membrane coat | 5/210 | 11/1126 | 2.437229 | 0.0376445 | 0.1158209 | 0.2128523 | 408418 410030 411147 412083 725057 | AP-2 complex subunit mu; protein SEC13 homolog; AP-2 complex subunit alpha; coatomer subunit gamma; coatomer subunit beta |
| Module 2 | GO: Cellular component | GO:0005615 | extracellular space | 4/210 | 8/1126 | 2.680952 | 0.0441243 | 0.1158209 | 0.2128523 | 406078 408937 413749 100577408 | transferrin 1; SPARC; ovalbumin-related protein X; leukocyte elastase inhibitor-like |
| Module 2 | GO: Cellular component | GO:0012506 | vesicle membrane | 3/210 | 5/1126 | 3.217143 | 0.0476910 | 0.1158209 | 0.2128523 | 410030 412083 725057 | protein SEC13 homolog; coatomer subunit gamma; coatomer subunit beta |
| Module 2 | GO: Cellular component | GO:0030660 | Golgi-associated vesicle membrane | 3/210 | 5/1126 | 3.217143 | 0.0476910 | 0.1158209 | 0.2128523 | 410030 412083 725057 | protein SEC13 homolog; coatomer subunit gamma; coatomer subunit beta |
| Module 2 | GO: Cellular component | GO:0030662 | coated vesicle membrane | 3/210 | 5/1126 | 3.217143 | 0.0476910 | 0.1158209 | 0.2128523 | 410030 412083 725057 | protein SEC13 homolog; coatomer subunit gamma; coatomer subunit beta |
| Module 3 | GO: Molecular function | GO:0038023 | signaling receptor activity | 18/117 | 43/1400 | 5.008945 | 0.0000000 | 0.0000000 | 0.0000001 | 406079 406082 406127 406153 408729 409042 409227 410478 410788 411212 411323 411738 412299 412740 413040 551848 552552 100576449 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; ultraviolet-sensitive opsin; nicotinic acetylcholine receptor alpha2 subunit; uncharacterized LOC408729; corazonin receptor; ultraspiracle; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like; discoidin domain-containing receptor 2-like; serotonin receptor; lutropin-choriogonadotropic hormone receptor; muscarinic acetylcholine receptor DM1; ligand-gated chloride channel homolog 3; probable G-protein coupled receptor 52; protocadherin-like wing polarity protein stan; uncharacterized LOC552552; protein patched |
| Module 3 | GO: Molecular function | GO:0022803 | passive transmembrane transporter activity | 12/117 | 19/1400 | 7.557355 | 0.0000000 | 0.0000000 | 0.0000001 | 406079 406082 406153 410478 410788 411732 412740 413020 413259 552552 725219 726724 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like; trimeric intracellular cation channel type B; ligand-gated chloride channel homolog 3; two pore potassium channel protein sup-9; neurogenic protein big brain; uncharacterized LOC552552; TWiK family of potassium channels protein 7-like; transient receptor potential channel pyrexia |
| Module 3 | GO: Cellular component | GO:0098794 | postsynapse | 6/111 | 6/1126 | 10.144144 | 0.0000008 | 0.0000154 | 0.0000199 | 406079 406082 406153 410478 410788 552552 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like; uncharacterized LOC552552 |
| Module 3 | GO: Cellular component | GO:0071944 | cell periphery | 13/111 | 31/1126 | 4.253996 | 0.0000019 | 0.0000180 | 0.0000349 | 406079 406082 406153 410368 410478 410788 411212 411744 412299 412740 413524 551848 552552 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; cadherin-23; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like; discoidin domain-containing receptor 2-like; spectrin beta chain; muscarinic acetylcholine receptor DM1; ligand-gated chloride channel homolog 3; exocyst complex component 1; protocadherin-like wing polarity protein stan; uncharacterized LOC552552 |
| Module 3 | GO: Cellular component | GO:0005886 | plasma membrane | 11/111 | 25/1126 | 4.463423 | 0.0000072 | 0.0000301 | 0.0000902 | 406079 406082 406153 410368 410478 410788 411212 412299 412740 551848 552552 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; cadherin-23; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like; discoidin domain-containing receptor 2-like; muscarinic acetylcholine receptor DM1; ligand-gated chloride channel homolog 3; protocadherin-like wing polarity protein stan; uncharacterized LOC552552 |
| Module 3 | GO: Cellular component | GO:0045211 | postsynaptic membrane | 5/111 | 5/1126 | 10.144144 | 0.0000086 | 0.0000301 | 0.0000902 | 406079 406082 406153 410478 410788 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like |
| Module 3 | GO: Cellular component | GO:0097060 | synaptic membrane | 5/111 | 5/1126 | 10.144144 | 0.0000086 | 0.0000301 | 0.0000902 | 406079 406082 406153 410478 410788 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like |
| Module 3 | GO: Cellular component | GO:0016021 | integral component of membrane | 67/111 | 461/1126 | 1.474312 | 0.0000111 | 0.0000301 | 0.0000909 | 406079 406082 406127 406153 408329 408500 408613 408729 408908 409042 409056 409087 409089 409139 409164 409511 409640 409691 409931 409957 410368 410478 410485 410613 410788 410902 411199 411212 411323 411732 411738 411750 412299 412465 412740 412965 413020 413030 413040 413133 413259 413689 413916 550899 551375 551765 551848 552139 552194 552477 552552 552612 552721 552735 724220 724552 724679 724761 725011 725219 726095 726322 726724 726941 100576355 100576449 100577325 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; ultraviolet-sensitive opsin; nicotinic acetylcholine receptor alpha2 subunit; uncharacterized LOC408329; endothelin-converting enzyme 1; synaptotagmin 1; uncharacterized LOC408729; anoctamin-8; corazonin receptor; sodium-dependent phosphate transporter 2; D-beta-hydroxybutyrate dehydrogenase, mitochondrial; vesicular inhibitory amino acid transporter; uncharacterized LOC409139; uncharacterized LOC409164; tetraspanin-1; proton-coupled amino acid transporter 1-like; signal peptide peptidase-like 3; protein FAM69C; synaptotagmin 4; cadherin-23; nicotinic acetylcholine receptor beta1 subunit; transmembrane and TPR repeat-containing protein CG4050-like; major facilitator superfamily domain-containing protein 6; glutamate receptor ionotropic, kainate 2-like; 18-wheeler; sodium-dependent neutral amino acid transporter B(0)AT3; discoidin domain-containing receptor 2-like; serotonin receptor; trimeric intracellular cation channel type B; lutropin-choriogonadotropic hormone receptor; probable E3 ubiquitin-protein ligase HERC4; muscarinic acetylcholine receptor DM1; tetraspanin-7; ligand-gated chloride channel homolog 3; synaptotagmin-14; two pore potassium channel protein sup-9; transmembrane protein 189; probable G-protein coupled receptor 52; heparan sulfate glucosamine 3-O-sulfotransferase 6; neurogenic protein big brain; solute carrier family 12 member 8; transmembrane protein 64; UPF0769 protein C21orf59 homolog; receptor-type tyrosine-protein phosphatase N2; uncharacterized LOC551765; protocadherin-like wing polarity protein stan; 1-acyl-sn-glycerol-3-phosphate acyltransferase alpha-like; transmembrane protein 181; dipeptidyl aminopeptidase-like protein 6; uncharacterized LOC552552; uncharacterized LOC552612; CD63 antigen; uncharacterized LOC552735; synaptotagmin-10; elongation of very long chain fatty acids protein AAEL008004-like; uncharacterized LOC724679; uncharacterized LOC724761; mitochondrial inner membrane protein COX18; TWiK family of potassium channels protein 7-like; galactoside 2-alpha-L-fucosyltransferase 2-like; uncharacterized LOC726322; transient receptor potential channel pyrexia; ATP-binding cassette sub-family A member 2-like; uncharacterized LOC100576355; protein patched; uncharacterized LOC100577325 |
| Module 3 | GO: Cellular component | GO:0031224 | intrinsic component of membrane | 67/111 | 461/1126 | 1.474312 | 0.0000111 | 0.0000301 | 0.0000909 | 406079 406082 406127 406153 408329 408500 408613 408729 408908 409042 409056 409087 409089 409139 409164 409511 409640 409691 409931 409957 410368 410478 410485 410613 410788 410902 411199 411212 411323 411732 411738 411750 412299 412465 412740 412965 413020 413030 413040 413133 413259 413689 413916 550899 551375 551765 551848 552139 552194 552477 552552 552612 552721 552735 724220 724552 724679 724761 725011 725219 726095 726322 726724 726941 100576355 100576449 100577325 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; ultraviolet-sensitive opsin; nicotinic acetylcholine receptor alpha2 subunit; uncharacterized LOC408329; endothelin-converting enzyme 1; synaptotagmin 1; uncharacterized LOC408729; anoctamin-8; corazonin receptor; sodium-dependent phosphate transporter 2; D-beta-hydroxybutyrate dehydrogenase, mitochondrial; vesicular inhibitory amino acid transporter; uncharacterized LOC409139; uncharacterized LOC409164; tetraspanin-1; proton-coupled amino acid transporter 1-like; signal peptide peptidase-like 3; protein FAM69C; synaptotagmin 4; cadherin-23; nicotinic acetylcholine receptor beta1 subunit; transmembrane and TPR repeat-containing protein CG4050-like; major facilitator superfamily domain-containing protein 6; glutamate receptor ionotropic, kainate 2-like; 18-wheeler; sodium-dependent neutral amino acid transporter B(0)AT3; discoidin domain-containing receptor 2-like; serotonin receptor; trimeric intracellular cation channel type B; lutropin-choriogonadotropic hormone receptor; probable E3 ubiquitin-protein ligase HERC4; muscarinic acetylcholine receptor DM1; tetraspanin-7; ligand-gated chloride channel homolog 3; synaptotagmin-14; two pore potassium channel protein sup-9; transmembrane protein 189; probable G-protein coupled receptor 52; heparan sulfate glucosamine 3-O-sulfotransferase 6; neurogenic protein big brain; solute carrier family 12 member 8; transmembrane protein 64; UPF0769 protein C21orf59 homolog; receptor-type tyrosine-protein phosphatase N2; uncharacterized LOC551765; protocadherin-like wing polarity protein stan; 1-acyl-sn-glycerol-3-phosphate acyltransferase alpha-like; transmembrane protein 181; dipeptidyl aminopeptidase-like protein 6; uncharacterized LOC552552; uncharacterized LOC552612; CD63 antigen; uncharacterized LOC552735; synaptotagmin-10; elongation of very long chain fatty acids protein AAEL008004-like; uncharacterized LOC724679; uncharacterized LOC724761; mitochondrial inner membrane protein COX18; TWiK family of potassium channels protein 7-like; galactoside 2-alpha-L-fucosyltransferase 2-like; uncharacterized LOC726322; transient receptor potential channel pyrexia; ATP-binding cassette sub-family A member 2-like; uncharacterized LOC100576355; protein patched; uncharacterized LOC100577325 |
| Module 3 | GO: Biological process | GO:0007165 | signal transduction | 18/60 | 106/960 | 2.716981 | 0.0000269 | 0.0012388 | 0.0023151 | 406083 406127 408500 408809 408830 408880 409113 409903 410206 410332 410630 410887 410902 413473 551848 724880 100126690 100576924 | tachykinin; ultraviolet-sensitive opsin; endothelin-converting enzyme 1; diacylglycerol kinase theta; muscle M-line assembly protein unc-89; guanylate cyclase, soluble, beta 1; guanine nucleotide-binding protein subunit beta-5; GTP-binding protein RAD-like; differentially expressed in FDCP 8 homolog; breast cancer anti-estrogen resistance protein 3; cGMP-specific 3’,5’-cyclic phosphodiesterase; myotubularin-related protein 13; 18-wheeler; GTPase-activating Rap/Ran-GAP domain-like protein 3; protocadherin-like wing polarity protein stan; allatostatin A; pheromone biosynthesis-activating neuropeptide; ankyrin repeat and death domain-containing protein 1A-like |
| Module 3 | GO: Cellular component | GO:0044459 | plasma membrane part | 7/111 | 12/1126 | 5.917417 | 0.0000394 | 0.0000936 | 0.0002904 | 406079 406082 406153 410478 410788 411212 412299 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like; discoidin domain-containing receptor 2-like; muscarinic acetylcholine receptor DM1 |
| Module 3 | KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 6/58 | 14/1081 | 7.987685 | 0.0000396 | 0.0017440 | 0.0019610 | 406079 409042 411323 412299 412740 552552 | NMDA receptor 1; corazonin receptor; serotonin receptor; muscarinic acetylcholine receptor DM1; ligand-gated chloride channel homolog 3; uncharacterized LOC552552 |
| Module 3 | GO: Cellular component | GO:0098590 | plasma membrane region | 5/111 | 6/1126 | 8.453454 | 0.0000474 | 0.0001000 | 0.0002910 | 406079 406082 406153 410478 410788 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like |
| Module 3 | GO: Molecular function | GO:0015318 | inorganic molecular entity transmembrane transporter activity | 13/117 | 46/1400 | 3.381642 | 0.0000478 | 0.0004943 | 0.0003843 | 406079 406082 406153 409056 410478 410788 411199 411732 412740 413020 552552 725219 726724 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; sodium-dependent phosphate transporter 2; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like; sodium-dependent neutral amino acid transporter B(0)AT3; trimeric intracellular cation channel type B; ligand-gated chloride channel homolog 3; two pore potassium channel protein sup-9; uncharacterized LOC552552; TWiK family of potassium channels protein 7-like; transient receptor potential channel pyrexia |
| Module 3 | GO: Biological process | GO:0007186 | G-protein coupled receptor signaling pathway | 5/60 | 9/960 | 8.888889 | 0.0000841 | 0.0019338 | 0.0054207 | 406083 408500 408809 724880 100126690 | tachykinin; endothelin-converting enzyme 1; diacylglycerol kinase theta; allatostatin A; pheromone biosynthesis-activating neuropeptide |
| Module 3 | GO: Biological process | GO:0050794 | regulation of cellular process | 27/60 | 228/960 | 1.894737 | 0.0001505 | 0.0023074 | 0.0077617 | 406083 406086 406127 408500 408809 408830 408880 409113 409903 410206 410332 410468 410630 410887 410902 411744 413473 551848 552145 724220 724301 724551 724880 726648 100126690 100576333 100576924 | tachykinin; dorsal; ultraviolet-sensitive opsin; endothelin-converting enzyme 1; diacylglycerol kinase theta; muscle M-line assembly protein unc-89; guanylate cyclase, soluble, beta 1; guanine nucleotide-binding protein subunit beta-5; GTP-binding protein RAD-like; differentially expressed in FDCP 8 homolog; breast cancer anti-estrogen resistance protein 3; protein hairy; cGMP-specific 3’,5’-cyclic phosphodiesterase; myotubularin-related protein 13; 18-wheeler; spectrin beta chain; GTPase-activating Rap/Ran-GAP domain-like protein 3; protocadherin-like wing polarity protein stan; repressor of RNA polymerase III transcription MAF1 homolog; synaptotagmin-10; paired mesoderm homeobox protein 2-like; BRCA1-associated RING domain protein 1-like; allatostatin A; fas apoptotic inhibitory molecule 1; pheromone biosynthesis-activating neuropeptide; nucleoredoxin-like; ankyrin repeat and death domain-containing protein 1A-like |
| Module 3 | GO: Molecular function | GO:0015075 | ion transmembrane transporter activity | 12/117 | 47/1400 | 3.055101 | 0.0002796 | 0.0021670 | 0.0020322 | 406079 406082 406153 410478 410788 411199 411732 412740 413020 552552 725219 726724 | NMDA receptor 1; nicotinic acetylcholine receptor alpha8 subunit; nicotinic acetylcholine receptor alpha2 subunit; nicotinic acetylcholine receptor beta1 subunit; glutamate receptor ionotropic, kainate 2-like; sodium-dependent neutral amino acid transporter B(0)AT3; trimeric intracellular cation channel type B; ligand-gated chloride channel homolog 3; two pore potassium channel protein sup-9; uncharacterized LOC552552; TWiK family of potassium channels protein 7-like; transient receptor potential channel pyrexia |
| Module 3 | GO: Biological process | GO:0012501 | programmed cell death | 3/60 | 5/960 | 9.600000 | 0.0021249 | 0.0244358 | 0.0456657 | 724551 724743 726648 | BRCA1-associated RING domain protein 1-like; PRKC apoptosis WT1 regulator protein-like; fas apoptotic inhibitory molecule 1 |
| Module 3 | KEGG | KEGG:04214 | Apoptosis - fly | 5/58 | 19/1081 | 4.904719 | 0.0024358 | 0.0535875 | 0.0602538 | 408758 409227 409523 413809 100576402 | ecdysteroid-regulated gene E74; ultraspiracle; mitogen-activated protein kinase 1; mitogen-activated protein kinase kinase kinase 7; serine/threonine-protein kinase atr-like |
| Module 3 | KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 4/58 | 17/1081 | 4.385395 | 0.0105173 | 0.1542536 | 0.1734431 | 408446 409155 410396 551958 | probable aconitate hydratase, mitochondrial; dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial-like; isocitrate dehydrogenase [NAD] subunit gamma, mitochondrial; uncharacterized LOC551958 |
| Module 3 | KEGG | KEGG:01200 | Carbon metabolism | 6/58 | 40/1081 | 2.795690 | 0.0169304 | 0.1862344 | 0.2094023 | 408446 409155 409682 410396 550785 551958 | probable aconitate hydratase, mitochondrial; dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial-like; NADP-dependent malic enzyme; isocitrate dehydrogenase [NAD] subunit gamma, mitochondrial; fructose-bisphosphate aldolase; uncharacterized LOC551958 |
| Module 3 | GO: Biological process | GO:0006099 | tricarboxylic acid cycle | 3/60 | 10/960 | 4.800000 | 0.0203347 | 0.1870790 | 0.3277629 | 408446 409155 410396 | probable aconitate hydratase, mitochondrial; dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial-like; isocitrate dehydrogenase [NAD] subunit gamma, mitochondrial |
| Module 3 | KEGG | KEGG:04310 | Wnt signaling pathway | 4/58 | 21/1081 | 3.550082 | 0.0225143 | 0.1981256 | 0.2227728 | 411738 413809 551313 724997 | lutropin-choriogonadotropic hormone receptor; mitogen-activated protein kinase kinase kinase 7; calcineurin subunit B type 2; protein naked cuticle homolog 2-like |
| Module 3 | GO: Biological process | GO:0006887 | exocytosis | 3/60 | 11/960 | 4.363636 | 0.0267324 | 0.2049482 | 0.3447069 | 412965 413524 724220 | synaptotagmin-14; exocyst complex component 1; synaptotagmin-10 |
| Module 3 | KEGG | KEGG:00350 | Tyrosine metabolism | 2/58 | 6/1081 | 6.212644 | 0.0369259 | 0.2707900 | 0.3044768 | 725400 751102 | 4-hydroxyphenylpyruvate dioxygenase; tyramine beta hydroxylase |
| Module 3 | GO: Biological process | GO:0035556 | intracellular signal transduction | 7/60 | 54/960 | 2.074074 | 0.0450967 | 0.2963496 | 0.4845915 | 408809 408830 408880 410206 410332 410887 413473 | diacylglycerol kinase theta; muscle M-line assembly protein unc-89; guanylate cyclase, soluble, beta 1; differentially expressed in FDCP 8 homolog; breast cancer anti-estrogen resistance protein 3; myotubularin-related protein 13; GTPase-activating Rap/Ran-GAP domain-like protein 3 |
| Module 4 | KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 22/132 | 58/1081 | 3.106322 | 0.0000002 | 0.0000113 | 0.0000127 | 408557 408706 409377 409587 409613 409911 409968 412045 412141 412505 412526 413441 413976 551559 551853 552295 552378 552747 724587 725571 726087 727262 | thioredoxin domain-containing protein 5; heat shock protein 70Cb ortholog; transitional endoplasmic reticulum ATPase TER94; heat shock 70 kDa protein cognate 3; GTP-binding protein SAR1; protein disulfide-isomerase A3; protein transport protein Sec31A; dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2; tumor suppressor candidate 3; translocation protein SEC63 homolog; protein disulfide-isomerase A6; dnaJ homolog subfamily C member 3; derlin-2; translocon-associated protein subunit beta; STT3, subunit of the oligosaccharyltransferase complex, homolog B; translocon-associated protein subunit gamma-like; membrane-associated ring finger (C3HC4) 6; glucosidase 2 subunit beta; nucleotide exchange factor SIL1; protein transport protein Sec61 subunit alpha; ribophorin I; derlin-1-like |
| Module 4 | GO: Biological process | GO:0006082 | organic acid metabolic process | 20/92 | 64/960 | 3.260870 | 0.0000004 | 0.0000196 | 0.0000178 | 409945 410422 411796 411923 412166 412670 412674 412675 412876 412948 550686 550828 551088 551103 551154 552014 552417 552712 724712 725031 | aspartate–tRNA ligase, cytoplasmic; dihydrofolate reductase; serine hydroxymethyltransferase; alanine–tRNA ligase, cytoplasmic; acyl-CoA Delta(11) desaturase; probable phosphoserine aminotransferase; phosphoserine phosphatase; aspartate aminotransferase, mitochondrial; pyruvate carboxylase, mitochondrial; delta-1-pyrroline-5-carboxylate synthase; ATP-citrate synthase; elongation of very long chain fatty acids protein AAEL008004-like; asparagine–tRNA ligase, cytoplasmic; probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial; glucose-6-phosphate isomerase; probable methylthioribulose-1-phosphate dehydratase; acyl-CoA Delta(11) desaturase; 6-phosphogluconate dehydrogenase, decarboxylating; serine–tRNA ligase, mitochondrial; elongation of very long chain fatty acids protein 6 |
| Module 4 | GO: Biological process | GO:0044283 | small molecule biosynthetic process | 14/92 | 40/960 | 3.652174 | 0.0000061 | 0.0001615 | 0.0002511 | 410422 410539 411796 412166 412670 412674 412876 412948 550828 551143 551154 552014 552417 725031 | dihydrofolate reductase; protein 5NUC-like; serine hydroxymethyltransferase; acyl-CoA Delta(11) desaturase; probable phosphoserine aminotransferase; phosphoserine phosphatase; pyruvate carboxylase, mitochondrial; delta-1-pyrroline-5-carboxylate synthase; elongation of very long chain fatty acids protein AAEL008004-like; inositol-3-phosphate synthase 1-B; glucose-6-phosphate isomerase; probable methylthioribulose-1-phosphate dehydratase; acyl-CoA Delta(11) desaturase; elongation of very long chain fatty acids protein 6 |
| Module 4 | KEGG | KEGG:01200 | Carbon metabolism | 15/132 | 40/1081 | 3.071023 | 0.0000254 | 0.0007364 | 0.0008286 | 409736 411796 412670 412674 412675 412876 413867 443552 550804 551103 551154 551276 552712 725325 100577053 | methylmalonate-semialdehyde dehydrogenase [acylating]-like protein; serine hydroxymethyltransferase; probable phosphoserine aminotransferase; phosphoserine phosphatase; aspartate aminotransferase, mitochondrial; pyruvate carboxylase, mitochondrial; transaldolase; catalase; transketolase; probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial; glucose-6-phosphate isomerase; isocitrate dehydrogenase [NADP] cytoplasmic; 6-phosphogluconate dehydrogenase, decarboxylating; glucose-6-phosphate 1-dehydrogenase; glycine dehydrogenase (decarboxylating), mitochondrial |
| Module 4 | KEGG | KEGG:01100 | Metabolic pathways | 59/132 | 321/1081 | 1.505216 | 0.0000686 | 0.0013270 | 0.0014932 | 406076 406114 408642 408817 408955 409515 409736 410422 410530 410539 410554 411662 411715 411796 411897 412045 412141 412663 412670 412674 412675 412815 412876 412889 412948 413228 413356 413601 413763 413867 413942 414008 550686 550687 550804 551103 551143 551154 551276 551837 551853 551872 552014 552148 552242 552286 552328 552712 725031 725146 725325 726087 726156 726818 726880 727078 727599 100577053 100577378 | vacuolar H+ ATP synthase 16 kDa proteolipid subunit; alpha-amylase; NAD kinase 2, mitochondrial-like; alanine–glyoxylate aminotransferase 2-like; 4-aminobutyrate aminotransferase, mitochondrial; long-chain-fatty-acid–CoA ligase 4; methylmalonate-semialdehyde dehydrogenase [acylating]-like protein; dihydrofolate reductase; alkaline phosphatase-like; protein 5NUC-like; methylcrotonoyl-CoA carboxylase beta chain, mitochondrial; probable trans-2-enoyl-CoA reductase, mitochondrial; phospholipase D3-like; serine hydroxymethyltransferase; phosphoglucomutase; dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2; tumor suppressor candidate 3; laminin subunit alpha; probable phosphoserine aminotransferase; phosphoserine phosphatase; aspartate aminotransferase, mitochondrial; fatty acid synthase; pyruvate carboxylase, mitochondrial; branched-chain-amino-acid aminotransferase, cytosolic-like; delta-1-pyrroline-5-carboxylate synthase; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; UDP-glucose 6-dehydrogenase; adenine phosphoribosyltransferase; hydroxymethylglutaryl-CoA synthase 1; transaldolase; acyl-CoA synthetase short-chain family member 3, mitochondrial; ribonucleoside-diphosphate reductase subunit M2; ATP-citrate synthase; aldehyde dehydrogenase, mitochondrial; transketolase; probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial; inositol-3-phosphate synthase 1-B; glucose-6-phosphate isomerase; isocitrate dehydrogenase [NADP] cytoplasmic; long-chain-fatty-acid–CoA ligase ACSBG2; STT3, subunit of the oligosaccharyltransferase complex, homolog B; porphobilinogen deaminase; probable methylthioribulose-1-phosphate dehydratase; thymidylate kinase; uncharacterized LOC552242; acetyl-CoA carboxylase; glycogen [starch] synthase; 6-phosphogluconate dehydrogenase, decarboxylating; elongation of very long chain fatty acids protein 6; very-long-chain enoyl-CoA reductase; glucose-6-phosphate 1-dehydrogenase; ribophorin I; pantothenate kinase 3; beta-hexosaminidase subunit beta-like; pancreatic lipase-related protein 2-like; phospholipid phosphatase 3-like; NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial; glycine dehydrogenase (decarboxylating), mitochondrial; galactokinase-like |
| Module 4 | GO: Cellular component | GO:0005783 | endoplasmic reticulum | 9/115 | 23/1126 | 3.831380 | 0.0002124 | 0.0053105 | 0.0190058 | 409264 409613 409911 411533 412505 413976 551559 552313 727262 | ATPase ASNA1 homolog; GTP-binding protein SAR1; protein disulfide-isomerase A3; alpha-2-macroglobulin receptor-associated protein; translocation protein SEC63 homolog; derlin-2; translocon-associated protein subunit beta; sterol O-acyltransferase 1; derlin-1-like |
| Module 4 | KEGG | KEGG:01212 | Fatty acid metabolism | 9/132 | 20/1081 | 3.685227 | 0.0002398 | 0.0034765 | 0.0039119 | 409515 411662 412166 412815 551837 552286 552417 725031 725146 | long-chain-fatty-acid–CoA ligase 4; probable trans-2-enoyl-CoA reductase, mitochondrial; acyl-CoA Delta(11) desaturase; fatty acid synthase; long-chain-fatty-acid–CoA ligase ACSBG2; acetyl-CoA carboxylase; acyl-CoA Delta(11) desaturase; elongation of very long chain fatty acids protein 6; very-long-chain enoyl-CoA reductase |
| Module 4 | GO: Biological process | GO:0051186 | cofactor metabolic process | 10/92 | 31/960 | 3.366059 | 0.0003279 | 0.0048104 | 0.0070759 | 408642 410422 411796 413228 550686 551103 551154 552014 552712 725325 | NAD kinase 2, mitochondrial-like; dihydrofolate reductase; serine hydroxymethyltransferase; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; ATP-citrate synthase; probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial; glucose-6-phosphate isomerase; probable methylthioribulose-1-phosphate dehydratase; 6-phosphogluconate dehydrogenase, decarboxylating; glucose-6-phosphate 1-dehydrogenase |
| Module 4 | KEGG | KEGG:01230 | Biosynthesis of amino acids | 10/132 | 25/1081 | 3.275758 | 0.0003445 | 0.0039960 | 0.0044964 | 411796 412670 412674 412675 412876 412889 412948 413867 550804 551276 | serine hydroxymethyltransferase; probable phosphoserine aminotransferase; phosphoserine phosphatase; aspartate aminotransferase, mitochondrial; pyruvate carboxylase, mitochondrial; branched-chain-amino-acid aminotransferase, cytosolic-like; delta-1-pyrroline-5-carboxylate synthase; transaldolase; transketolase; isocitrate dehydrogenase [NADP] cytoplasmic |
| Module 4 | GO: Biological process | GO:0006520 | cellular amino acid metabolic process | 11/92 | 37/960 | 3.102233 | 0.0003631 | 0.0048104 | 0.0070759 | 409945 410422 411796 411923 412670 412674 412675 412948 551088 552014 724712 | aspartate–tRNA ligase, cytoplasmic; dihydrofolate reductase; serine hydroxymethyltransferase; alanine–tRNA ligase, cytoplasmic; probable phosphoserine aminotransferase; phosphoserine phosphatase; aspartate aminotransferase, mitochondrial; delta-1-pyrroline-5-carboxylate synthase; asparagine–tRNA ligase, cytoplasmic; probable methylthioribulose-1-phosphate dehydratase; serine–tRNA ligase, mitochondrial |
| Module 4 | GO: Biological process | GO:0005975 | carbohydrate metabolic process | 11/92 | 38/960 | 3.020595 | 0.0004707 | 0.0049896 | 0.0084854 | 406114 411100 411897 412876 551143 551154 552328 552712 725325 726445 100577378 | alpha-amylase; FGGY carbohydrate kinase domain-containing protein; phosphoglucomutase; pyruvate carboxylase, mitochondrial; inositol-3-phosphate synthase 1-B; glucose-6-phosphate isomerase; glycogen [starch] synthase; 6-phosphogluconate dehydrogenase, decarboxylating; glucose-6-phosphate 1-dehydrogenase; glycerol-3-phosphate dehydrogenase; galactokinase-like |
| Module 4 | GO: Molecular function | GO:0019842 | vitamin binding | 7/144 | 16/1400 | 4.253472 | 0.0005444 | 0.0168693 | 0.0720431 | 408817 411796 412675 412815 412876 413228 724919 | alanine–glyoxylate aminotransferase 2-like; serine hydroxymethyltransferase; aspartate aminotransferase, mitochondrial; fatty acid synthase; pyruvate carboxylase, mitochondrial; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; mitochondrial amidoxime reducing component 2-like |
| Module 4 | GO: Biological process | GO:0006629 | lipid metabolic process | 10/92 | 34/960 | 3.069054 | 0.0007581 | 0.0066966 | 0.0128619 | 411706 412166 413763 550686 550828 551143 552417 725031 725146 100576414 | phospholipase B1, membrane-associated-like; acyl-CoA Delta(11) desaturase; hydroxymethylglutaryl-CoA synthase 1; ATP-citrate synthase; elongation of very long chain fatty acids protein AAEL008004-like; inositol-3-phosphate synthase 1-B; acyl-CoA Delta(11) desaturase; elongation of very long chain fatty acids protein 6; very-long-chain enoyl-CoA reductase; putative fatty acyl-CoA reductase CG5065 |
| Module 4 | GO: Molecular function | GO:0016903 | oxidoreductase activity, acting on the aldehyde or oxo group of donors | 5/144 | 9/1400 | 5.401235 | 0.0009640 | 0.0168693 | 0.0720431 | 409736 412948 550687 551103 100576414 | methylmalonate-semialdehyde dehydrogenase [acylating]-like protein; delta-1-pyrroline-5-carboxylate synthase; aldehyde dehydrogenase, mitochondrial; probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial; putative fatty acyl-CoA reductase CG5065 |
| Module 4 | KEGG | KEGG:00030 | Pentose phosphate pathway | 6/132 | 12/1081 | 4.094697 | 0.0014704 | 0.0142134 | 0.0159933 | 411897 413867 550804 551154 552712 725325 | phosphoglucomutase; transaldolase; transketolase; glucose-6-phosphate isomerase; 6-phosphogluconate dehydrogenase, decarboxylating; glucose-6-phosphate 1-dehydrogenase |
| Module 4 | KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 4/132 | 6/1081 | 5.459596 | 0.0026240 | 0.0217419 | 0.0244645 | 412166 552417 725031 725146 | acyl-CoA Delta(11) desaturase; acyl-CoA Delta(11) desaturase; elongation of very long chain fatty acids protein 6; very-long-chain enoyl-CoA reductase |
| Module 4 | GO: Molecular function | GO:0050662 | coenzyme binding | 11/144 | 44/1400 | 2.430556 | 0.0035707 | 0.0366561 | 0.0987647 | 408817 410422 411796 412212 412675 412876 413228 413356 724919 725325 726445 | alanine–glyoxylate aminotransferase 2-like; dihydrofolate reductase; serine hydroxymethyltransferase; apoptosis-inducing factor 1, mitochondrial; aspartate aminotransferase, mitochondrial; pyruvate carboxylase, mitochondrial; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; UDP-glucose 6-dehydrogenase; mitochondrial amidoxime reducing component 2-like; glucose-6-phosphate 1-dehydrogenase; glycerol-3-phosphate dehydrogenase |
| Module 4 | GO: Molecular function | GO:0016746 | transferase activity, transferring acyl groups | 8/144 | 27/1400 | 2.880658 | 0.0041893 | 0.0366561 | 0.0987647 | 409905 412815 413228 413763 550686 550828 552313 725031 | 2-acylglycerol O-acyltransferase 1-like; fatty acid synthase; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; hydroxymethylglutaryl-CoA synthase 1; ATP-citrate synthase; elongation of very long chain fatty acids protein AAEL008004-like; sterol O-acyltransferase 1; elongation of very long chain fatty acids protein 6 |
| Module 4 | KEGG | KEGG:00061 | Fatty acid biosynthesis | 4/132 | 7/1081 | 4.679654 | 0.0055448 | 0.0357335 | 0.0402083 | 409515 412815 551837 552286 | long-chain-fatty-acid–CoA ligase 4; fatty acid synthase; long-chain-fatty-acid–CoA ligase ACSBG2; acetyl-CoA carboxylase |
| Module 4 | KEGG | KEGG:04512 | ECM-receptor interaction | 4/132 | 7/1081 | 4.679654 | 0.0055448 | 0.0357335 | 0.0402083 | 408551 408552 412663 726736 | collagen alpha-5(IV) chain; collagen alpha-1(IV) chain; laminin subunit alpha; laminin subunit beta-1 |
| Module 4 | GO: Molecular function | GO:0070279 | vitamin B6 binding | 5/144 | 13/1400 | 3.739316 | 0.0069904 | 0.0489331 | 0.1160987 | 408817 411796 412675 413228 724919 | alanine–glyoxylate aminotransferase 2-like; serine hydroxymethyltransferase; aspartate aminotransferase, mitochondrial; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; mitochondrial amidoxime reducing component 2-like |
| Module 4 | GO: Biological process | GO:0055086 | nucleobase-containing small molecule metabolic process | 11/92 | 52/960 | 2.207358 | 0.0074910 | 0.0567177 | 0.0583937 | 408642 410539 413601 414008 551154 552014 552148 552316 552511 552712 725325 | NAD kinase 2, mitochondrial-like; protein 5NUC-like; adenine phosphoribosyltransferase; ribonucleoside-diphosphate reductase subunit M2; glucose-6-phosphate isomerase; probable methylthioribulose-1-phosphate dehydratase; thymidylate kinase; GMP reductase 1-like; GTP:AMP phosphotransferase AK3, mitochondrial; 6-phosphogluconate dehydrogenase, decarboxylating; glucose-6-phosphate 1-dehydrogenase |
| Module 4 | KEGG | KEGG:03050 | Proteasome | 8/132 | 26/1081 | 2.519814 | 0.0091992 | 0.0529757 | 0.0596097 | 409168 409609 409880 410095 411206 413757 551128 551550 | 26S proteasome non-ATPase regulatory subunit 13; 26S proteasome non-ATPase regulatory subunit 4; 26S proteasome non-ATPase regulatory subunit 12; proteasome subunit alpha type-7-1; proteasome subunit beta type-5-like; uncharacterized LOC413757; 26S protease regulatory subunit 4; probable 26S proteasome non-ATPase regulatory subunit 3 |
| Module 4 | KEGG | KEGG:00500 | Starch and sucrose metabolism | 4/132 | 8/1081 | 4.094697 | 0.0100471 | 0.0529757 | 0.0596097 | 406114 411897 551154 552328 | alpha-amylase; phosphoglucomutase; glucose-6-phosphate isomerase; glycogen [starch] synthase |
| Module 4 | GO: Biological process | GO:0005996 | monosaccharide metabolic process | 4/92 | 10/960 | 4.173913 | 0.0105758 | 0.0700644 | 0.0726256 | 412876 551154 725325 100577378 | pyruvate carboxylase, mitochondrial; glucose-6-phosphate isomerase; glucose-6-phosphate 1-dehydrogenase; galactokinase-like |
| Module 4 | GO: Cellular component | GO:0005789 | endoplasmic reticulum membrane | 5/115 | 15/1126 | 3.263768 | 0.0132381 | 0.0690393 | 0.1840125 | 412505 413976 551559 552313 727262 | translocation protein SEC63 homolog; derlin-2; translocon-associated protein subunit beta; sterol O-acyltransferase 1; derlin-1-like |
| Module 4 | GO: Cellular component | GO:0044432 | endoplasmic reticulum part | 5/115 | 15/1126 | 3.263768 | 0.0132381 | 0.0690393 | 0.1840125 | 412505 413976 551559 552313 727262 | translocation protein SEC63 homolog; derlin-2; translocon-associated protein subunit beta; sterol O-acyltransferase 1; derlin-1-like |
| Module 4 | GO: Biological process | GO:0044262 | cellular carbohydrate metabolic process | 3/92 | 6/960 | 5.217391 | 0.0137676 | 0.0810759 | 0.0902469 | 551143 552328 552712 | inositol-3-phosphate synthase 1-B; glycogen [starch] synthase; 6-phosphogluconate dehydrogenase, decarboxylating |
| Module 4 | KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 5/132 | 13/1081 | 3.149767 | 0.0143762 | 0.0660047 | 0.0742702 | 411796 412670 412674 413228 100577053 | serine hydroxymethyltransferase; probable phosphoserine aminotransferase; phosphoserine phosphatase; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; glycine dehydrogenase (decarboxylating), mitochondrial |
| Module 4 | KEGG | KEGG:00565 | Ether lipid metabolism | 3/132 | 5/1081 | 4.913636 | 0.0147942 | 0.0660047 | 0.0742702 | 411715 552242 727078 | phospholipase D3-like; uncharacterized LOC552242; phospholipid phosphatase 3-like |
| Module 4 | GO: Molecular function | GO:0017171 | serine hydrolase activity | 6/144 | 21/1400 | 2.777778 | 0.0156822 | 0.0914793 | 0.1953392 | 409162 410451 411889 411896 413645 726126 | retinoid-inducible serine carboxypeptidase-like; venom serine carboxypeptidase; putative serine protease K12H4.7; prolyl endopeptidase-like; trypsin alpha-3; proclotting enzyme |
| Module 4 | GO: Biological process | GO:1901576 | organic substance biosynthetic process | 37/92 | 286/960 | 1.349954 | 0.0161832 | 0.0850415 | 0.1017507 | 408443 408642 408981 409799 409928 409945 410422 410539 411510 411796 411923 412166 412670 412674 412876 412948 413228 413601 413763 414008 550686 550828 551088 551103 551143 551154 551872 552014 552148 552328 552417 552511 552642 552764 724712 725031 100577998 | uncharacterized LOC408443; NAD kinase 2, mitochondrial-like; activating transcription factor 3; ataxin-7-like protein 3; RNA polymerase II elongation factor Ell; aspartate–tRNA ligase, cytoplasmic; dihydrofolate reductase; protein 5NUC-like; DNA-directed RNA polymerases I and III subunit RPAC1; serine hydroxymethyltransferase; alanine–tRNA ligase, cytoplasmic; acyl-CoA Delta(11) desaturase; probable phosphoserine aminotransferase; phosphoserine phosphatase; pyruvate carboxylase, mitochondrial; delta-1-pyrroline-5-carboxylate synthase; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; adenine phosphoribosyltransferase; hydroxymethylglutaryl-CoA synthase 1; ribonucleoside-diphosphate reductase subunit M2; ATP-citrate synthase; elongation of very long chain fatty acids protein AAEL008004-like; asparagine–tRNA ligase, cytoplasmic; probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial; inositol-3-phosphate synthase 1-B; glucose-6-phosphate isomerase; porphobilinogen deaminase; probable methylthioribulose-1-phosphate dehydratase; thymidylate kinase; glycogen [starch] synthase; acyl-CoA Delta(11) desaturase; GTP:AMP phosphotransferase AK3, mitochondrial; DNA replication licensing factor Mcm3; eukaryotic translation initiation factor 2A; serine–tRNA ligase, mitochondrial; elongation of very long chain fatty acids protein 6; origin recognition complex subunit 3 |
| Module 4 | KEGG | KEGG:03060 | Protein export | 4/132 | 9/1081 | 3.639731 | 0.0163914 | 0.0679073 | 0.0764111 | 409587 412505 552114 725571 | heat shock 70 kDa protein cognate 3; translocation protein SEC63 homolog; signal recognition particle 54 kDa protein; protein transport protein Sec61 subunit alpha |
| Module 4 | GO: Cellular component | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network | 5/115 | 16/1126 | 3.059783 | 0.0177131 | 0.0690393 | 0.1840125 | 412505 413976 551559 552313 727262 | translocation protein SEC63 homolog; derlin-2; translocon-associated protein subunit beta; sterol O-acyltransferase 1; derlin-1-like |
| Module 4 | GO: Molecular function | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | 5/144 | 16/1400 | 3.038194 | 0.0183912 | 0.0919562 | 0.2114621 | 412815 413356 552712 725325 726445 | fatty acid synthase; UDP-glucose 6-dehydrogenase; 6-phosphogluconate dehydrogenase, decarboxylating; glucose-6-phosphate 1-dehydrogenase; glycerol-3-phosphate dehydrogenase |
| Module 4 | GO: Biological process | GO:0006793 | phosphorus metabolic process | 13/92 | 74/960 | 1.833138 | 0.0184013 | 0.0850415 | 0.1105693 | 408642 410539 414008 550686 551103 551143 551154 552148 552316 552511 552712 725325 726445 | NAD kinase 2, mitochondrial-like; protein 5NUC-like; ribonucleoside-diphosphate reductase subunit M2; ATP-citrate synthase; probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial; inositol-3-phosphate synthase 1-B; glucose-6-phosphate isomerase; thymidylate kinase; GMP reductase 1-like; GTP:AMP phosphotransferase AK3, mitochondrial; 6-phosphogluconate dehydrogenase, decarboxylating; glucose-6-phosphate 1-dehydrogenase; glycerol-3-phosphate dehydrogenase |
| Module 4 | GO: Cellular component | GO:0016021 | integral component of membrane | 58/115 | 461/1126 | 1.231878 | 0.0190797 | 0.0690393 | 0.1840125 | 406076 408523 409041 409905 410429 410530 410844 410878 411105 411266 411902 412094 412166 412212 412431 412505 413000 413117 413189 413840 413976 550691 550828 550886 550914 551036 551165 551180 551217 551324 551388 551559 551795 551844 551936 552256 552295 552313 552401 552417 552746 552747 552769 724497 724919 724952 725031 725146 725245 725324 725420 725571 726987 727078 727262 100576236 100576414 100577231 | vacuolar H+ ATP synthase 16 kDa proteolipid subunit; protein TRC8 homolog; transmembrane protein 115; 2-acylglycerol O-acyltransferase 1-like; mannose-P-dolichol utilization defect 1 protein homolog; alkaline phosphatase-like; neuronal membrane glycoprotein M6-a; epoxide hydrolase 4-like; luciferin 4-monooxygenase; nicalin; transmembrane protein 205; protein catecholamines up; acyl-CoA Delta(11) desaturase; apoptosis-inducing factor 1, mitochondrial; thiamine transporter 2-like; translocation protein SEC63 homolog; proton-coupled amino acid transporter 1; proton-coupled amino acid transporter 1; NADH-cytochrome b5 reductase 2; glucose-6-phosphate exchanger SLC37A2; derlin-2; putative inorganic phosphate cotransporter; elongation of very long chain fatty acids protein AAEL008004-like; atlastin; DNA ligase 1-like; vacuole membrane protein 1; innexin inx3; puromycin-sensitive aminopeptidase-like protein; high affinity copper uptake protein 1; transmembrane protein 19; adipokinetic hormone receptor; translocon-associated protein subunit beta; xenotropic and polytropic retrovirus receptor 1 homolog; stromal cell-derived factor 2-like; facilitated trehalose transporter Tret1; peroxisomal membrane protein PEX14; translocon-associated protein subunit gamma-like; sterol O-acyltransferase 1; protein cueball; acyl-CoA Delta(11) desaturase; 17-beta-hydroxysteroid dehydrogenase 13-like; glucosidase 2 subunit beta; ubiA prenyltransferase domain-containing protein 1 homolog; patched domain-containing protein 3-like; mitochondrial amidoxime reducing component 2-like; sodium-independent sulfate anion transporter-like; elongation of very long chain fatty acids protein 6; very-long-chain enoyl-CoA reductase; protein eiger-like; uncharacterized LOC725324; sodium-dependent nutrient amino acid transporter 1-like; protein transport protein Sec61 subunit alpha; uncharacterized LOC726987; phospholipid phosphatase 3-like; derlin-1-like; uncharacterized LOC100576236; putative fatty acyl-CoA reductase CG5065; uncharacterized LOC100577231 |
| Module 4 | GO: Cellular component | GO:0031224 | intrinsic component of membrane | 58/115 | 461/1126 | 1.231878 | 0.0190797 | 0.0690393 | 0.1840125 | 406076 408523 409041 409905 410429 410530 410844 410878 411105 411266 411902 412094 412166 412212 412431 412505 413000 413117 413189 413840 413976 550691 550828 550886 550914 551036 551165 551180 551217 551324 551388 551559 551795 551844 551936 552256 552295 552313 552401 552417 552746 552747 552769 724497 724919 724952 725031 725146 725245 725324 725420 725571 726987 727078 727262 100576236 100576414 100577231 | vacuolar H+ ATP synthase 16 kDa proteolipid subunit; protein TRC8 homolog; transmembrane protein 115; 2-acylglycerol O-acyltransferase 1-like; mannose-P-dolichol utilization defect 1 protein homolog; alkaline phosphatase-like; neuronal membrane glycoprotein M6-a; epoxide hydrolase 4-like; luciferin 4-monooxygenase; nicalin; transmembrane protein 205; protein catecholamines up; acyl-CoA Delta(11) desaturase; apoptosis-inducing factor 1, mitochondrial; thiamine transporter 2-like; translocation protein SEC63 homolog; proton-coupled amino acid transporter 1; proton-coupled amino acid transporter 1; NADH-cytochrome b5 reductase 2; glucose-6-phosphate exchanger SLC37A2; derlin-2; putative inorganic phosphate cotransporter; elongation of very long chain fatty acids protein AAEL008004-like; atlastin; DNA ligase 1-like; vacuole membrane protein 1; innexin inx3; puromycin-sensitive aminopeptidase-like protein; high affinity copper uptake protein 1; transmembrane protein 19; adipokinetic hormone receptor; translocon-associated protein subunit beta; xenotropic and polytropic retrovirus receptor 1 homolog; stromal cell-derived factor 2-like; facilitated trehalose transporter Tret1; peroxisomal membrane protein PEX14; translocon-associated protein subunit gamma-like; sterol O-acyltransferase 1; protein cueball; acyl-CoA Delta(11) desaturase; 17-beta-hydroxysteroid dehydrogenase 13-like; glucosidase 2 subunit beta; ubiA prenyltransferase domain-containing protein 1 homolog; patched domain-containing protein 3-like; mitochondrial amidoxime reducing component 2-like; sodium-independent sulfate anion transporter-like; elongation of very long chain fatty acids protein 6; very-long-chain enoyl-CoA reductase; protein eiger-like; uncharacterized LOC725324; sodium-dependent nutrient amino acid transporter 1-like; protein transport protein Sec61 subunit alpha; uncharacterized LOC726987; phospholipid phosphatase 3-like; derlin-1-like; uncharacterized LOC100576236; putative fatty acyl-CoA reductase CG5065; uncharacterized LOC100577231 |
| Module 4 | GO: Cellular component | GO:0000502 | proteasome complex | 4/115 | 11/1126 | 3.560474 | 0.0193310 | 0.0690393 | 0.1840125 | 410095 411206 413757 551550 | proteasome subunit alpha type-7-1; proteasome subunit beta type-5-like; uncharacterized LOC413757; probable 26S proteasome non-ATPase regulatory subunit 3 |
| Module 4 | KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 6/132 | 19/1081 | 2.586124 | 0.0208348 | 0.0805614 | 0.0906498 | 411897 413189 413356 551154 726818 100577378 | phosphoglucomutase; NADH-cytochrome b5 reductase 2; UDP-glucose 6-dehydrogenase; glucose-6-phosphate isomerase; beta-hexosaminidase subunit beta-like; galactokinase-like |
| Module 4 | GO: Biological process | GO:0019637 | organophosphate metabolic process | 11/92 | 60/960 | 1.913043 | 0.0220585 | 0.0850415 | 0.1168592 | 408642 410539 414008 551143 551154 552148 552316 552511 552712 725325 726445 | NAD kinase 2, mitochondrial-like; protein 5NUC-like; ribonucleoside-diphosphate reductase subunit M2; inositol-3-phosphate synthase 1-B; glucose-6-phosphate isomerase; thymidylate kinase; GMP reductase 1-like; GTP:AMP phosphotransferase AK3, mitochondrial; 6-phosphogluconate dehydrogenase, decarboxylating; glucose-6-phosphate 1-dehydrogenase; glycerol-3-phosphate dehydrogenase |
| Module 4 | GO: Biological process | GO:0006575 | cellular modified amino acid metabolic process | 3/92 | 7/960 | 4.472050 | 0.0224344 | 0.0850415 | 0.1168592 | 410422 411796 552014 | dihydrofolate reductase; serine hydroxymethyltransferase; probable methylthioribulose-1-phosphate dehydratase |
| Module 4 | GO: Biological process | GO:0044249 | cellular biosynthetic process | 36/92 | 282/960 | 1.332100 | 0.0224638 | 0.0850415 | 0.1168592 | 408443 408642 408981 409799 409928 409945 410422 410539 411510 411796 411923 412166 412670 412674 412948 413228 413601 413763 414008 550686 550828 551088 551103 551143 551154 551872 552014 552148 552328 552417 552511 552642 552764 724712 725031 100577998 | uncharacterized LOC408443; NAD kinase 2, mitochondrial-like; activating transcription factor 3; ataxin-7-like protein 3; RNA polymerase II elongation factor Ell; aspartate–tRNA ligase, cytoplasmic; dihydrofolate reductase; protein 5NUC-like; DNA-directed RNA polymerases I and III subunit RPAC1; serine hydroxymethyltransferase; alanine–tRNA ligase, cytoplasmic; acyl-CoA Delta(11) desaturase; probable phosphoserine aminotransferase; phosphoserine phosphatase; delta-1-pyrroline-5-carboxylate synthase; 5-aminolevulinate synthase, erythroid-specific, mitochondrial; adenine phosphoribosyltransferase; hydroxymethylglutaryl-CoA synthase 1; ribonucleoside-diphosphate reductase subunit M2; ATP-citrate synthase; elongation of very long chain fatty acids protein AAEL008004-like; asparagine–tRNA ligase, cytoplasmic; probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial; inositol-3-phosphate synthase 1-B; glucose-6-phosphate isomerase; porphobilinogen deaminase; probable methylthioribulose-1-phosphate dehydratase; thymidylate kinase; glycogen [starch] synthase; acyl-CoA Delta(11) desaturase; GTP:AMP phosphotransferase AK3, mitochondrial; DNA replication licensing factor Mcm3; eukaryotic translation initiation factor 2A; serine–tRNA ligase, mitochondrial; elongation of very long chain fatty acids protein 6; origin recognition complex subunit 3 |
| Module 4 | GO: Cellular component | GO:1905368 | peptidase complex | 5/115 | 17/1126 | 2.879795 | 0.0230885 | 0.0721516 | 0.1878012 | 409799 410095 411206 413757 551550 | ataxin-7-like protein 3; proteasome subunit alpha type-7-1; proteasome subunit beta type-5-like; uncharacterized LOC413757; probable 26S proteasome non-ATPase regulatory subunit 3 |
| Module 4 | GO: Biological process | GO:0044255 | cellular lipid metabolic process | 6/92 | 25/960 | 2.504348 | 0.0259888 | 0.0918273 | 0.1315040 | 412166 413763 550828 551143 552417 725031 | acyl-CoA Delta(11) desaturase; hydroxymethylglutaryl-CoA synthase 1; elongation of very long chain fatty acids protein AAEL008004-like; inositol-3-phosphate synthase 1-B; acyl-CoA Delta(11) desaturase; elongation of very long chain fatty acids protein 6 |
| Module 4 | KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 6/132 | 21/1081 | 2.339827 | 0.0339122 | 0.1204778 | 0.1355648 | 408955 409736 410554 412889 413763 550687 | 4-aminobutyrate aminotransferase, mitochondrial; methylmalonate-semialdehyde dehydrogenase [acylating]-like protein; methylcrotonoyl-CoA carboxylase beta chain, mitochondrial; branched-chain-amino-acid aminotransferase, cytosolic-like; hydroxymethylglutaryl-CoA synthase 1; aldehyde dehydrogenase, mitochondrial |
| Module 4 | KEGG | KEGG:00510 | N-Glycan biosynthesis | 4/132 | 11/1081 | 2.977961 | 0.0353124 | 0.1204778 | 0.1355648 | 412045 412141 551853 726087 | dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2; tumor suppressor candidate 3; STT3, subunit of the oligosaccharyltransferase complex, homolog B; ribophorin I |
| Module 4 | GO: Molecular function | GO:0016875 | ligase activity, forming carbon-oxygen bonds | 4/144 | 13/1400 | 2.991453 | 0.0366761 | 0.1604580 | 0.3364074 | 409945 411923 551088 724712 | aspartate–tRNA ligase, cytoplasmic; alanine–tRNA ligase, cytoplasmic; asparagine–tRNA ligase, cytoplasmic; serine–tRNA ligase, mitochondrial |
| Module 4 | KEGG | KEGG:00062 | Fatty acid elongation | 3/132 | 7/1081 | 3.509740 | 0.0430289 | 0.1386486 | 0.1560111 | 411662 725031 725146 | probable trans-2-enoyl-CoA reductase, mitochondrial; elongation of very long chain fatty acids protein 6; very-long-chain enoyl-CoA reductase |
| Module 4 | GO: Cellular component | GO:0012505 | endomembrane system | 11/115 | 63/1126 | 1.709593 | 0.0479692 | 0.1332479 | 0.3576653 | 409264 409613 409911 411533 412505 413976 551559 552313 552346 724795 727262 | ATPase ASNA1 homolog; GTP-binding protein SAR1; protein disulfide-isomerase A3; alpha-2-macroglobulin receptor-associated protein; translocation protein SEC63 homolog; derlin-2; translocon-associated protein subunit beta; sterol O-acyltransferase 1; coatomer subunit delta; WAS protein family homolog 1-like; derlin-1-like |
| Module 5 | GO: Biological process | GO:0046907 | intracellular transport | 8/44 | 60/960 | 2.909091 | 0.0043197 | 0.0462494 | 0.0745260 | 408391 410493 413344 413614 413845 551140 552830 726833 | AP-1 complex subunit mu-1; vacuolar protein sorting-associated protein 11 homolog; exportin-5; ras-related protein Rab-14; mitochondrial import receptor subunit TOM20 homolog; signal transducing adapter molecule 1; alpha-soluble NSF attachment protein; TOM1-like protein 2 |
| Module 5 | KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 7/50 | 47/1081 | 3.220000 | 0.0044720 | 0.1477696 | 0.1675121 | 409225 409607 410565 410838 411182 551174 551511 | NEDD8-conjugating enzyme Ubc12; ubiquitin-conjugating enzyme E2 Q2; cullin-1; ubiquitin-conjugating enzyme E2 R2; ubiquitin-protein ligase E3A; E3 SUMO-protein ligase PIAS3; transcription elongation factor B polypeptide 2 |
| Module 5 | GO: Biological process | GO:0070727 | cellular macromolecule localization | 7/44 | 49/960 | 3.116883 | 0.0052370 | 0.0462494 | 0.0745260 | 408391 410493 413344 413845 551140 552830 726833 | AP-1 complex subunit mu-1; vacuolar protein sorting-associated protein 11 homolog; exportin-5; mitochondrial import receptor subunit TOM20 homolog; signal transducing adapter molecule 1; alpha-soluble NSF attachment protein; TOM1-like protein 2 |
| Module 5 | GO: Biological process | GO:0051649 | establishment of localization in cell | 8/44 | 62/960 | 2.815249 | 0.0053229 | 0.0462494 | 0.0745260 | 408391 410493 413344 413614 413845 551140 552830 726833 | AP-1 complex subunit mu-1; vacuolar protein sorting-associated protein 11 homolog; exportin-5; ras-related protein Rab-14; mitochondrial import receptor subunit TOM20 homolog; signal transducing adapter molecule 1; alpha-soluble NSF attachment protein; TOM1-like protein 2 |
| Module 5 | KEGG | KEGG:04142 | Lysosome | 5/50 | 28/1081 | 3.860714 | 0.0075779 | 0.1477696 | 0.1675121 | 406108 408391 551012 552375 725899 | tetraspanin 6; AP-1 complex subunit mu-1; cation-independent mannose-6-phosphate receptor; Niemann-Pick C1 protein; alpha-N-acetylgalactosaminidase |
| Module 5 | GO: Biological process | GO:0032535 | regulation of cellular component size | 3/44 | 10/960 | 6.545454 | 0.0086108 | 0.0462494 | 0.0745260 | 409218 409581 552486 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Biological process | GO:0032970 | regulation of actin filament-based process | 3/44 | 10/960 | 6.545454 | 0.0086108 | 0.0462494 | 0.0745260 | 409218 409581 552486 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Biological process | GO:0043254 | regulation of protein complex assembly | 3/44 | 10/960 | 6.545454 | 0.0086108 | 0.0462494 | 0.0745260 | 409218 409581 552486 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Biological process | GO:0044087 | regulation of cellular component biogenesis | 3/44 | 10/960 | 6.545454 | 0.0086108 | 0.0462494 | 0.0745260 | 409218 409581 552486 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Biological process | GO:0090066 | regulation of anatomical structure size | 3/44 | 10/960 | 6.545454 | 0.0086108 | 0.0462494 | 0.0745260 | 409218 409581 552486 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Molecular function | GO:0004721 | phosphoprotein phosphatase activity | 4/65 | 19/1400 | 4.534413 | 0.0096915 | 0.1506384 | 0.2560559 | 408701 409430 550710 551854 | protein phosphatase 1H; serine/threonine-protein phosphatase alpha-2 isoform; serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform; serine/threonine-protein phosphatase 5 |
| Module 5 | GO: Biological process | GO:0045184 | establishment of protein localization | 8/44 | 69/960 | 2.529644 | 0.0103044 | 0.0462494 | 0.0859993 | 408391 410493 413344 413845 551140 552830 726833 100576745 | AP-1 complex subunit mu-1; vacuolar protein sorting-associated protein 11 homolog; exportin-5; mitochondrial import receptor subunit TOM20 homolog; signal transducing adapter molecule 1; alpha-soluble NSF attachment protein; TOM1-like protein 2; leucine-rich repeat-containing protein 26-like |
| Module 5 | GO: Biological process | GO:0042592 | homeostatic process | 4/44 | 20/960 | 4.363636 | 0.0108764 | 0.0462494 | 0.0862321 | 411391 552501 726237 726649 | thioredoxin-related transmembrane protein 2 homolog; transmembrane and coiled-coil domain-containing protein 1; protein disulfide-isomerase TMX3; thioredoxin domain-containing protein 15 |
| Module 5 | GO: Biological process | GO:0008104 | protein localization | 8/44 | 70/960 | 2.493507 | 0.0112337 | 0.0462494 | 0.0862321 | 408391 410493 413344 413845 551140 552830 726833 100576745 | AP-1 complex subunit mu-1; vacuolar protein sorting-associated protein 11 homolog; exportin-5; mitochondrial import receptor subunit TOM20 homolog; signal transducing adapter molecule 1; alpha-soluble NSF attachment protein; TOM1-like protein 2; leucine-rich repeat-containing protein 26-like |
| Module 5 | GO: Biological process | GO:0006810 | transport | 13/44 | 149/960 | 1.903600 | 0.0118084 | 0.0462494 | 0.0862321 | 408391 408828 409476 410493 412806 413344 413614 413845 551140 552830 726431 726833 100576745 | AP-1 complex subunit mu-1; uncharacterized MFS-type transporter C09D4.1; nuclear cap-binding protein subunit 1; vacuolar protein sorting-associated protein 11 homolog; vesicle transport protein GOT1B; exportin-5; ras-related protein Rab-14; mitochondrial import receptor subunit TOM20 homolog; signal transducing adapter molecule 1; alpha-soluble NSF attachment protein; uncharacterized LOC726431; TOM1-like protein 2; leucine-rich repeat-containing protein 26-like |
| Module 5 | KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 3/50 | 12/1081 | 5.405000 | 0.0152561 | 0.1983298 | 0.2248273 | 413430 413742 551554 | RAC serine/threonine-protein kinase; signal transducer and activator of transcription 5B; ras-related protein Rac1 |
| Module 5 | GO: Biological process | GO:0097435 | supramolecular fiber organization | 3/44 | 13/960 | 5.034965 | 0.0186288 | 0.0589261 | 0.0955373 | 409218 409581 552486 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Biological process | GO:0022411 | cellular component disassembly | 2/44 | 5/960 | 8.727273 | 0.0188062 | 0.0589261 | 0.0955373 | 409581 552486 | F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Biological process | GO:0051129 | negative regulation of cellular component organization | 2/44 | 5/960 | 8.727273 | 0.0188062 | 0.0589261 | 0.0955373 | 409581 552486 | F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Molecular function | GO:0070279 | vitamin B6 binding | 3/65 | 13/1400 | 4.970414 | 0.0195674 | 0.1506384 | 0.2560559 | 409196 409927 410638 | alanine aminotransferase 1; putative pyridoxal-dependent decarboxylase domain-containing protein 2; aromatic-L-amino-acid decarboxylase |
| Module 5 | GO: Biological process | GO:0030036 | actin cytoskeleton organization | 3/44 | 14/960 | 4.675325 | 0.0229589 | 0.0674419 | 0.1117738 | 409218 409581 552486 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Biological process | GO:0045454 | cell redox homeostasis | 3/44 | 15/960 | 4.363636 | 0.0277918 | 0.0725676 | 0.1273434 | 411391 726237 726649 | thioredoxin-related transmembrane protein 2 homolog; protein disulfide-isomerase TMX3; thioredoxin domain-containing protein 15 |
| Module 5 | GO: Biological process | GO:0051128 | regulation of cellular component organization | 3/44 | 15/960 | 4.363636 | 0.0277918 | 0.0725676 | 0.1273434 | 409218 409581 552486 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha |
| Module 5 | GO: Molecular function | GO:0016830 | carbon-carbon lyase activity | 2/65 | 6/1400 | 7.179487 | 0.0282176 | 0.1506384 | 0.2560559 | 409927 410638 | putative pyridoxal-dependent decarboxylase domain-containing protein 2; aromatic-L-amino-acid decarboxylase |
| Module 5 | GO: Molecular function | GO:0008092 | cytoskeletal protein binding | 4/65 | 26/1400 | 3.313610 | 0.0292798 | 0.1506384 | 0.2560559 | 409218 409581 552486 725218 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; F-actin-capping protein subunit alpha; growth arrest-specific protein 2-like |
| Module 5 | GO: Molecular function | GO:0016788 | hydrolase activity, acting on ester bonds | 6/65 | 52/1400 | 2.485207 | 0.0300237 | 0.1506384 | 0.2560559 | 408701 408840 409430 413429 550710 551854 | protein phosphatase 1H; 5’-3’ exoribonuclease 2 homolog; serine/threonine-protein phosphatase alpha-2 isoform; acid phosphatase type 7; serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform; serine/threonine-protein phosphatase 5 |
| Module 5 | KEGG | KEGG:04144 | Endocytosis | 6/50 | 54/1081 | 2.402222 | 0.0339182 | 0.3307020 | 0.3748849 | 409218 409581 410241 551012 551140 552486 | neural Wiskott-Aldrich syndrome protein; F-actin-capping protein subunit beta; ras-related protein Rab-10; cation-independent mannose-6-phosphate receptor; signal transducing adapter molecule 1; F-actin-capping protein subunit alpha |
| Module 5 | GO: Molecular function | GO:0019842 | vitamin binding | 3/65 | 16/1400 | 4.038462 | 0.0346866 | 0.1506384 | 0.2560559 | 409196 409927 410638 | alanine aminotransferase 1; putative pyridoxal-dependent decarboxylase domain-containing protein 2; aromatic-L-amino-acid decarboxylase |
| Module 5 | GO: Cellular component | GO:0016021 | integral component of membrane | 34/65 | 461/1126 | 1.277624 | 0.0375885 | 0.4322677 | 0.9957147 | 406108 408303 408751 408828 409136 409145 409263 409310 409976 410507 410825 411391 411408 412024 412291 412636 412806 413164 413740 413845 550918 551466 551678 551890 552067 552098 552501 724286 724585 725316 727001 100576745 100578046 100578526 | tetraspanin 6; protein lifeguard 1; E3 ubiquitin-protein ligase RNF185-like; uncharacterized MFS-type transporter C09D4.1; 72 kDa inositol polyphosphate 5-phosphatase; zinc transporter ZIP9; glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1; CSC1-like protein 2; transmembrane emp24 domain-containing protein 5; sodium-independent sulfate anion transporter-like; leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 3; thioredoxin-related transmembrane protein 2 homolog; CDP-diacylglycerol–inositol 3-phosphatidyltransferase; probable palmitoyltransferase ZDHHC16; uncharacterized protein KIAA2013 homolog; protein ABHD13; vesicle transport protein GOT1B; membrane-associated progesterone receptor component 1; iodotyrosine deiodinase 1; mitochondrial import receptor subunit TOM20 homolog; ATP-binding cassette sub-family G member 4; CAAX prenyl protease 1 homolog; receptor expression-enhancing protein 5-like; probable serine incorporator; uncharacterized LOC552067; syntaxin-8; transmembrane and coiled-coil domain-containing protein 1; uncharacterized LOC724286; uncharacterized LOC724585; transmembrane protein 208; solute carrier family 35 member C2-like; leucine-rich repeat-containing protein 26-like; probable G-protein coupled receptor Mth-like 1; solute carrier family 35 member E2-like |
| Module 5 | GO: Cellular component | GO:0031224 | intrinsic component of membrane | 34/65 | 461/1126 | 1.277624 | 0.0375885 | 0.4322677 | 0.9957147 | 406108 408303 408751 408828 409136 409145 409263 409310 409976 410507 410825 411391 411408 412024 412291 412636 412806 413164 413740 413845 550918 551466 551678 551890 552067 552098 552501 724286 724585 725316 727001 100576745 100578046 100578526 | tetraspanin 6; protein lifeguard 1; E3 ubiquitin-protein ligase RNF185-like; uncharacterized MFS-type transporter C09D4.1; 72 kDa inositol polyphosphate 5-phosphatase; zinc transporter ZIP9; glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1; CSC1-like protein 2; transmembrane emp24 domain-containing protein 5; sodium-independent sulfate anion transporter-like; leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 3; thioredoxin-related transmembrane protein 2 homolog; CDP-diacylglycerol–inositol 3-phosphatidyltransferase; probable palmitoyltransferase ZDHHC16; uncharacterized protein KIAA2013 homolog; protein ABHD13; vesicle transport protein GOT1B; membrane-associated progesterone receptor component 1; iodotyrosine deiodinase 1; mitochondrial import receptor subunit TOM20 homolog; ATP-binding cassette sub-family G member 4; CAAX prenyl protease 1 homolog; receptor expression-enhancing protein 5-like; probable serine incorporator; uncharacterized LOC552067; syntaxin-8; transmembrane and coiled-coil domain-containing protein 1; uncharacterized LOC724286; uncharacterized LOC724585; transmembrane protein 208; solute carrier family 35 member C2-like; leucine-rich repeat-containing protein 26-like; probable G-protein coupled receptor Mth-like 1; solute carrier family 35 member E2-like |
| Module 5 | GO: Molecular function | GO:0019787 | ubiquitin-like protein transferase activity | 3/65 | 17/1400 | 3.800905 | 0.0407516 | 0.1506384 | 0.2560559 | 411182 413468 551174 | ubiquitin-protein ligase E3A; protein ariadne-1; E3 SUMO-protein ligase PIAS3 |
| Module 5 | GO: Molecular function | GO:0001882 | nucleoside binding | 6/65 | 57/1400 | 2.267206 | 0.0446336 | 0.1506384 | 0.2560559 | 410241 411085 411704 413614 551554 724676 | ras-related protein Rab-10; ADP-ribosylation factor-like protein 2; guanine nucleotide-binding protein G(i) subunit alpha; ras-related protein Rab-14; ras-related protein Rac1; ADP-ribosylation factor-like protein 1 |
| Module 5 | KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 2/50 | 8/1081 | 5.405000 | 0.0491201 | 0.3831368 | 0.4343252 | 409196 410482 | alanine aminotransferase 1; adenylosuccinate lyase |
| Module 6 | GO: Molecular function | GO:0001882 | nucleoside binding | 9/57 | 57/1400 | 3.878116 | 0.0003174 | 0.0082528 | 0.0038424 | 409529 410906 410969 550723 551185 551731 552196 552448 724873 | ras-like protein 2; guanine nucleotide-binding protein subunit alpha homolog; ras-related protein Rab-9A; ras-related protein Rab-39B; GTP-binding protein 1; rho GTPase-activating protein 190; ras-related protein Rab-43; ADP-ribosylation factor-like protein 8B-A; ras-related protein Rab-7a |
| Module 6 | GO: Molecular function | GO:0016817 | hydrolase activity, acting on acid anhydrides | 10/57 | 107/1400 | 2.295458 | 0.0090480 | 0.1176243 | 0.0730191 | 409529 410463 410676 410906 410969 550723 551185 551731 552196 724873 | ras-like protein 2; ATPase WRNIP1-like; adenylate kinase isoenzyme 6; guanine nucleotide-binding protein subunit alpha homolog; ras-related protein Rab-9A; ras-related protein Rab-39B; GTP-binding protein 1; rho GTPase-activating protein 190; ras-related protein Rab-43; ras-related protein Rab-7a |
| Module 6 | KEGG | KEGG:03410 | Base excision repair | 2/33 | 9/1081 | 7.279461 | 0.0284604 | 0.4603898 | 0.5139918 | 725801 100577027 | G/T mismatch-specific thymine DNA glycosylase-like; DNA repair protein XRCC1 |
| Module 6 | GO: Biological process | GO:0007275 | multicellular organism development | 2/34 | 8/960 | 7.058823 | 0.0298325 | 0.5328338 | 0.6676729 | 410808 412008 | E3 ubiquitin-protein ligase Siah1; sprouty-related, EVH1 domain-containing protein 1 |
| Module 6 | KEGG | KEGG:04140 | Autophagy - animal | 4/33 | 40/1081 | 3.275758 | 0.0302736 | 0.4603898 | 0.5139918 | 408358 409529 409925 724873 | RB1-inducible coiled-coil protein 1; ras-like protein 2; dual specificity mitogen-activated protein kinase kinase dSOR1; ras-related protein Rab-7a |
| Module 6 | GO: Molecular function | GO:0004540 | ribonuclease activity | 2/57 | 8/1400 | 6.140351 | 0.0389514 | 0.2712372 | 0.2357583 | 100576272 100576289 | tRNA-splicing endonuclease subunit Sen34; uncharacterized LOC100576289 |
| Module 6 | GO: Molecular function | GO:0003676 | nucleic acid binding | 19/57 | 319/1400 | 1.462905 | 0.0417288 | 0.2712372 | 0.2377121 | 408678 410463 412545 413515 551421 551877 724212 725326 725512 726215 727285 100576272 100576289 100576411 100576565 100576908 100577027 100577561 100577696 | OTU domain-containing protein 7B-like; ATPase WRNIP1-like; zinc finger protein 277; zinc finger protein 62-like; tRNA selenocysteine 1-associated protein 1-like; putative high mobility group protein B1-like 1; 60 kDa SS-A/Ro ribonucleoprotein-like; tRNA pseudouridine synthase-like 1; RISC-loading complex subunit TARBP2; uncharacterized LOC726215; zinc finger protein 227-like; tRNA-splicing endonuclease subunit Sen34; uncharacterized LOC100576289; uncharacterized LOC100576411; zinc finger protein 511; polycomb protein Asx; DNA repair protein XRCC1; uncharacterized LOC100577561; fez family zinc finger protein 2-like |
| Module 6 | KEGG | KEGG:04350 | TGF-beta signaling pathway | 2/33 | 11/1081 | 5.955923 | 0.0418536 | 0.4603898 | 0.5139918 | 410776 412866 | protein 60A; E3 ubiquitin-protein ligase SMURF2 |
| Module 7 | GO: Biological process | GO:0035556 | intracellular signal transduction | 6/23 | 54/960 | 4.637681 | 0.0011519 | 0.0380135 | 0.2050427 | 408373 412143 413034 413071 413448 413772 | phosphatidylinositol 4-kinase beta; rho guanine nucleotide exchange factor 18; rho-related BTB domain-containing protein 1; eye-specific diacylglycerol kinase; raf homolog serine/threonine-protein kinase phl; suppressor of cytokine signaling 7 |
| Module 7 | KEGG | KEGG:04068 | FoxO signaling pathway | 4/35 | 21/1081 | 5.882993 | 0.0037303 | 0.0687688 | 0.0701261 | 408438 413183 413448 552793 | insulin receptor substrate 1-B; G1/S-specific cyclin-D2; raf homolog serine/threonine-protein kinase phl; F-box only protein 32 |
| Module 7 | KEGG | KEGG:04320 | Dorso-ventral axis formation | 3/35 | 11/1081 | 8.423377 | 0.0042981 | 0.0687688 | 0.0701261 | 413448 727422 100577214 | raf homolog serine/threonine-protein kinase phl; beta-1,4-mannosyltransferase egh; protein giant-lens |
| Module 7 | KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 3/35 | 14/1081 | 6.618367 | 0.0088670 | 0.0766655 | 0.0781787 | 406070 406151 552518 | dopamine receptor 2; metabotropic glutamate receptor 1; serotonin receptor |
| Module 7 | KEGG | KEGG:00511 | Other glycan degradation | 2/35 | 5/1081 | 12.354286 | 0.0095832 | 0.0766655 | 0.0781787 | 412838 725756 | alpha-mannosidase 2; beta-galactosidase-like |
| Module 7 | KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 3/35 | 19/1081 | 4.876692 | 0.0211183 | 0.1295980 | 0.1321559 | 408373 413071 724991 | phosphatidylinositol 4-kinase beta; eye-specific diacylglycerol kinase; phosphatidylinositol 4-phosphate 5-kinase type-1 alpha |
| Module 7 | KEGG | KEGG:00562 | Inositol phosphate metabolism | 3/35 | 20/1081 | 4.632857 | 0.0242996 | 0.1295980 | 0.1321559 | 408373 724991 100577002 | phosphatidylinositol 4-kinase beta; phosphatidylinositol 4-phosphate 5-kinase type-1 alpha; multiple inositol polyphosphate phosphatase 1-like |
| Module 7 | GO: Biological process | GO:0007165 | signal transduction | 6/23 | 106/960 | 2.362592 | 0.0330865 | 0.5459279 | 0.7498662 | 408373 412143 413034 413071 413448 413772 | phosphatidylinositol 4-kinase beta; rho guanine nucleotide exchange factor 18; rho-related BTB domain-containing protein 1; eye-specific diacylglycerol kinase; raf homolog serine/threonine-protein kinase phl; suppressor of cytokine signaling 7 |
| Module 8 | KEGG | KEGG:00190 | Oxidative phosphorylation | 26/29 | 47/1081 | 20.620690 | 0.0000000 | 0.0000000 | 0.0000000 | 408367 408477 409114 409236 409473 409549 409930 411183 411411 412328 412396 413014 413605 551078 551660 551757 551766 551866 552424 552682 552699 724264 724719 725712 725797 726042 | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mitochondrial; ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; putative ATP synthase subunit f, mitochondrial; cytochrome b-c1 complex subunit Rieske, mitochondrial; NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial; NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial; cytochrome c oxidase subunit 4 isoform 1, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; cytochrome c1, heme protein, mitochondrial; NADH-quinone oxidoreductase subunit I; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial; uncharacterized LOC725712; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | KEGG | KEGG:01100 | Metabolic pathways | 27/29 | 321/1081 | 3.135353 | 0.0000000 | 0.0000000 | 0.0000000 | 408367 408477 409114 409236 409299 409473 409549 409930 411183 411411 412328 412396 413014 413605 551078 551660 551757 551766 551866 552424 552682 552699 724264 724719 725712 725797 726042 | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mitochondrial; ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; putative ATP synthase subunit f, mitochondrial; cytochrome b-c1 complex subunit Rieske, mitochondrial; NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial; NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial; cytochrome c oxidase subunit 4 isoform 1, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; cytochrome c1, heme protein, mitochondrial; NADH-quinone oxidoreductase subunit I; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial; uncharacterized LOC725712; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Biological process | GO:0017144 | drug metabolic process | 10/13 | 44/960 | 16.783217 | 0.0000000 | 0.0000000 | 0.0000000 | 409114 409236 409299 409473 409549 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Cellular component | GO:0044429 | mitochondrial part | 10/24 | 38/1126 | 12.346491 | 0.0000000 | 0.0000000 | 0.0000000 | 406075 408270 409473 410022 413014 551325 551757 724264 725797 726042 | ADP/ATP translocase; mitochondrial cytochrome C; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; mitochondrial-processing peptidase subunit beta; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; voltage-dependent anion-selective channel; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Biological process | GO:0055086 | nucleobase-containing small molecule metabolic process | 9/13 | 52/960 | 12.781065 | 0.0000000 | 0.0000000 | 0.0000000 | 409114 409236 409299 409473 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Cellular component | GO:0070469 | respiratory chain | 6/24 | 8/1126 | 35.187500 | 0.0000000 | 0.0000000 | 0.0000000 | 408270 409473 409549 551757 725797 726042 | mitochondrial cytochrome C; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Biological process | GO:0019637 | organophosphate metabolic process | 9/13 | 60/960 | 11.076923 | 0.0000000 | 0.0000000 | 0.0000000 | 409114 409236 409299 409473 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Cellular component | GO:0031967 | organelle envelope | 9/24 | 35/1126 | 12.064286 | 0.0000000 | 0.0000001 | 0.0000000 | 406075 408270 409473 413014 551325 551757 724264 725797 726042 | ADP/ATP translocase; mitochondrial cytochrome C; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; voltage-dependent anion-selective channel; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Cellular component | GO:0031975 | envelope | 9/24 | 35/1126 | 12.064286 | 0.0000000 | 0.0000001 | 0.0000000 | 406075 408270 409473 413014 551325 551757 724264 725797 726042 | ADP/ATP translocase; mitochondrial cytochrome C; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; voltage-dependent anion-selective channel; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Biological process | GO:1901135 | carbohydrate derivative metabolic process | 9/13 | 67/960 | 9.919633 | 0.0000000 | 0.0000001 | 0.0000000 | 409114 409236 409299 409473 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Biological process | GO:0022900 | electron transport chain | 5/13 | 9/960 | 41.025641 | 0.0000000 | 0.0000001 | 0.0000000 | 409473 413014 551757 724264 725797 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 |
| Module 8 | GO: Cellular component | GO:0031966 | mitochondrial membrane | 8/24 | 28/1126 | 13.404762 | 0.0000000 | 0.0000001 | 0.0000001 | 406075 409473 413014 551325 551757 724264 725797 726042 | ADP/ATP translocase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; voltage-dependent anion-selective channel; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Molecular function | GO:0009055 | electron transfer activity | 5/22 | 8/1400 | 39.772727 | 0.0000000 | 0.0000005 | 0.0000002 | 408270 409549 412396 413605 726042 | mitochondrial cytochrome C; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; cytochrome c oxidase subunit 4 isoform 1, mitochondrial; cytochrome c1, heme protein, mitochondrial; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Biological process | GO:0006793 | phosphorus metabolic process | 9/13 | 74/960 | 8.981289 | 0.0000000 | 0.0000001 | 0.0000000 | 409114 409236 409299 409473 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Cellular component | GO:0098803 | respiratory chain complex | 5/24 | 7/1126 | 33.511905 | 0.0000001 | 0.0000002 | 0.0000001 | 409473 409549 551757 725797 726042 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Biological process | GO:0006091 | generation of precursor metabolites and energy | 6/13 | 21/960 | 21.098901 | 0.0000001 | 0.0000003 | 0.0000001 | 409473 409549 413014 551757 724264 725797 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 |
| Module 8 | GO: Cellular component | GO:0098796 | membrane protein complex | 9/24 | 46/1126 | 9.179348 | 0.0000001 | 0.0000004 | 0.0000002 | 409114 409473 409549 551757 551766 552682 552699 725797 726042 | ATP synthase subunit alpha, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Molecular function | GO:0016651 | oxidoreductase activity, acting on NAD(P)H | 5/22 | 10/1400 | 31.818182 | 0.0000001 | 0.0000008 | 0.0000007 | 411411 413014 551078 551660 724264 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; NADH-quinone oxidoreductase subunit I; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Molecular function | GO:0015318 | inorganic molecular entity transmembrane transporter activity | 8/22 | 46/1400 | 11.067194 | 0.0000002 | 0.0000008 | 0.0000008 | 409114 409236 412396 551325 551766 552682 552699 726042 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; cytochrome c oxidase subunit 4 isoform 1, mitochondrial; voltage-dependent anion-selective channel; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Molecular function | GO:0015075 | ion transmembrane transporter activity | 8/22 | 47/1400 | 10.831722 | 0.0000002 | 0.0000008 | 0.0000008 | 409114 409236 412396 551325 551766 552682 552699 726042 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; cytochrome c oxidase subunit 4 isoform 1, mitochondrial; voltage-dependent anion-selective channel; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Cellular component | GO:0019866 | organelle inner membrane | 7/24 | 24/1126 | 13.684028 | 0.0000002 | 0.0000006 | 0.0000003 | 406075 409473 413014 551757 724264 725797 726042 | ADP/ATP translocase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Cellular component | GO:0045259 | proton-transporting ATP synthase complex | 4/24 | 5/1126 | 37.533333 | 0.0000008 | 0.0000021 | 0.0000010 | 409114 551766 552682 552699 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| Module 8 | GO: Biological process | GO:0015980 | energy derivation by oxidation of organic compounds | 5/13 | 17/960 | 21.719457 | 0.0000011 | 0.0000033 | 0.0000009 | 409473 409549 413014 551757 724264 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Cellular component | GO:0005746 | mitochondrial respiratory chain | 4/24 | 6/1126 | 31.277778 | 0.0000023 | 0.0000056 | 0.0000026 | 409473 551757 725797 726042 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Cellular component | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain | 4/24 | 7/1126 | 26.809524 | 0.0000053 | 0.0000107 | 0.0000053 | 409114 551766 552682 552699 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| Module 8 | GO: Cellular component | GO:0098800 | inner mitochondrial membrane protein complex | 4/24 | 7/1126 | 26.809524 | 0.0000053 | 0.0000107 | 0.0000053 | 409473 551757 725797 726042 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Molecular function | GO:0022804 | active transmembrane transporter activity | 5/22 | 19/1400 | 16.746412 | 0.0000060 | 0.0000203 | 0.0000146 | 409114 409236 551766 552682 552699 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| Module 8 | GO: Cellular component | GO:0031090 | organelle membrane | 8/24 | 55/1126 | 6.824242 | 0.0000078 | 0.0000144 | 0.0000072 | 406075 409473 413014 551325 551757 724264 725797 726042 | ADP/ATP translocase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; voltage-dependent anion-selective channel; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Cellular component | GO:1990204 | oxidoreductase complex | 4/24 | 8/1126 | 23.458333 | 0.0000105 | 0.0000181 | 0.0000092 | 409473 409549 551757 725797 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; succinate dehydrogenase cytochrome b560 subunit, mitochondrial; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 |
| Module 8 | GO: Cellular component | GO:0016469 | proton-transporting two-sector ATPase complex | 4/24 | 12/1126 | 15.638889 | 0.0000704 | 0.0001056 | 0.0000556 | 409114 551766 552682 552699 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| Module 8 | GO: Cellular component | GO:0044455 | mitochondrial membrane part | 4/24 | 12/1126 | 15.638889 | 0.0000704 | 0.0001056 | 0.0000556 | 409473 551757 725797 726042 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Biological process | GO:1901564 | organonitrogen compound metabolic process | 10/13 | 271/960 | 2.724950 | 0.0003445 | 0.0009187 | 0.0002497 | 409114 409236 409299 409473 410022 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; mitochondrial-processing peptidase subunit beta; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Cellular component | GO:0044444 | cytoplasmic part | 11/24 | 214/1126 | 2.411604 | 0.0022229 | 0.0031383 | 0.0016714 | 406075 408270 409473 410022 413014 551325 551660 551757 724264 725797 726042 | ADP/ATP translocase; mitochondrial cytochrome C; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; mitochondrial-processing peptidase subunit beta; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; voltage-dependent anion-selective channel; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Biological process | GO:0006139 | nucleobase-containing compound metabolic process | 9/13 | 285/960 | 2.331984 | 0.0035080 | 0.0084193 | 0.0024618 | 409114 409236 409299 409473 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Biological process | GO:0046483 | heterocycle metabolic process | 9/13 | 295/960 | 2.252934 | 0.0045674 | 0.0095806 | 0.0031066 | 409114 409236 409299 409473 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Biological process | GO:0006725 | cellular aromatic compound metabolic process | 9/13 | 297/960 | 2.237762 | 0.0048084 | 0.0095806 | 0.0032209 | 409114 409236 409299 409473 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Biological process | GO:1901360 | organic cyclic compound metabolic process | 9/13 | 300/960 | 2.215385 | 0.0051895 | 0.0095806 | 0.0033740 | 409114 409236 409299 409473 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 8 | GO: Molecular function | GO:0020037 | heme binding | 2/22 | 9/1400 | 14.141414 | 0.0079419 | 0.0192875 | 0.0163854 | 408270 413605 | mitochondrial cytochrome C; cytochrome c1, heme protein, mitochondrial |
| Module 8 | GO: Molecular function | GO:0046906 | tetrapyrrole binding | 2/22 | 9/1400 | 14.141414 | 0.0079419 | 0.0192875 | 0.0163854 | 408270 413605 | mitochondrial cytochrome C; cytochrome c1, heme protein, mitochondrial |
| Module 8 | GO: Biological process | GO:0006810 | transport | 6/13 | 149/960 | 2.973671 | 0.0084648 | 0.0145111 | 0.0051977 | 409114 409236 410022 551766 552682 552699 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; mitochondrial-processing peptidase subunit beta; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| Module 8 | GO: Cellular component | GO:0044446 | intracellular organelle part | 10/24 | 225/1126 | 2.085185 | 0.0116915 | 0.0155886 | 0.0083910 | 406075 408270 409473 410022 413014 551325 551757 724264 725797 726042 | ADP/ATP translocase; mitochondrial cytochrome C; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; mitochondrial-processing peptidase subunit beta; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; voltage-dependent anion-selective channel; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Cellular component | GO:0005737 | cytoplasm | 12/24 | 309/1126 | 1.822007 | 0.0146548 | 0.0185113 | 0.0096413 | 406075 408270 409299 409473 410022 413014 551325 551660 551757 724264 725797 726042 | ADP/ATP translocase; mitochondrial cytochrome C; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; mitochondrial-processing peptidase subunit beta; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5; voltage-dependent anion-selective channel; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial; cytochrome b-c1 complex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like; NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3; cytochrome c oxidase subunit 6A1, mitochondrial |
| Module 8 | GO: Molecular function | GO:0016817 | hydrolase activity, acting on acid anhydrides | 5/22 | 107/1400 | 2.973662 | 0.0218747 | 0.0464837 | 0.0389059 | 409114 409236 551766 552682 552699 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| Module 8 | GO: Biological process | GO:0034641 | cellular nitrogen compound metabolic process | 9/13 | 368/960 | 1.806020 | 0.0231719 | 0.0370750 | 0.0136592 | 409114 409236 409299 409473 551757 551766 552682 552699 724264 | ATP synthase subunit alpha, mitochondrial; ATP synthase subunit O, mitochondrial; adenylosuccinate synthetase; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8; cytochrome b-c1 complex subunit 7-like; ATP synthase subunit beta, mitochondrial; ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit; ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like |
| Module 9 | GO: Molecular function | GO:0020037 | heme binding | 2/17 | 9/1400 | 18.300654 | 0.0047547 | 0.0523021 | 0.0725722 | 408879 551179 | soluble guanylyl cyclase alpha 1 subunit; methyl farnesoate epoxidase |
| Module 9 | GO: Molecular function | GO:0046906 | tetrapyrrole binding | 2/17 | 9/1400 | 18.300654 | 0.0047547 | 0.0523021 | 0.0725722 | 408879 551179 | soluble guanylyl cyclase alpha 1 subunit; methyl farnesoate epoxidase |
| Module 9 | GO: Biological process | GO:0035556 | intracellular signal transduction | 3/12 | 54/960 | 4.444444 | 0.0257948 | 0.6240421 | 0.8680508 | 408879 411134 724571 | soluble guanylyl cyclase alpha 1 subunit; cdc42 homolog; atrial natriuretic peptide receptor 1 |
| Module 9 | GO: Molecular function | GO:0004672 | protein kinase activity | 3/17 | 57/1400 | 4.334365 | 0.0289984 | 0.1770952 | 0.3276403 | 408325 409908 724571 | cyclin-dependent kinase 5; tyrosine-protein kinase CSK; atrial natriuretic peptide receptor 1 |
| Module 9 | GO: Molecular function | GO:0016772 | transferase activity, transferring phosphorus-containing groups | 4/17 | 104/1400 | 3.167421 | 0.0321991 | 0.1770952 | 0.3276403 | 408325 409908 411191 724571 | cyclin-dependent kinase 5; tyrosine-protein kinase CSK; DNA-directed RNA polymerase III subunit RPC8; atrial natriuretic peptide receptor 1 |
| Module 9 | KEGG | KEGG:00230 | Purine metabolism | 2/10 | 35/1081 | 6.177143 | 0.0389432 | 0.1525425 | 0.1605711 | 408879 724571 | soluble guanylyl cyclase alpha 1 subunit; atrial natriuretic peptide receptor 1 |
Inspect the gene list for each module
This section lists all the genes in each module, their within-module connectivity values, and their fold-change in expression in response to queen pheromone in each of the four species.
Module 0
Table S25: List of all the genes in Module 0, ranked by their within-module connectivity, \(k\). The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
module_gene_list <- function(module){
inspect.module.genes(module) %>%
mutate(am_fc = log2(am_fc), bt_fc = log2(bt_fc), lf_fc = log2(lf_fc), ln_fc = log2(ln_fc)) %>%
rename(Gene = gene, Name = name)
}
gene_list <- module_gene_list(0)
saveRDS(gene_list, file = "supplement/tab_S25.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB40730 | odorant receptor 2 | 0.6572887 | 0.5427109 | -0.1048169 | 1.0856981 | -0.2756124 |
| GB52877 | uncharacterized protein LOC726699 isoform X2 | 0.5852105 | -0.5960765 | -0.1117507 | 0.2453807 | -0.4518517 |
| GB55921 | esterase FE4-like | 0.5261953 | -0.0722254 | -0.2349972 | 0.8469903 | -0.1497396 |
| GB42205 | uncharacterized protein LOC724413 | 0.5102071 | 0.4571923 | 0.0927301 | 0.2587316 | -0.1252817 |
| GB55743 | WD repeat-containing protein 78-like isoform X2 | 0.4763912 | -0.1199448 | 1.3331501 | 1.6458692 | -0.3359528 |
| GB43902 | hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like | 0.4071128 | -0.0658685 | -0.0051531 | 7.5139785 | -0.3675137 |
| GB54281 | cAMP-dependent protein kinase type II regulatory subunit isoform X2 | 0.3984861 | 0.0156292 | 0.0166301 | 0.1711831 | 0.1568573 |
| GB43306 | uncharacterized protein LOC100188904 | 0.3906286 | -0.7930497 | -0.2279818 | 0.9211482 | -0.2428097 |
| GB49904 | bone morphogenetic protein 5-like | 0.3796728 | -0.7719506 | 0.1965885 | 0.6831029 | -0.4370223 |
| GB45633 | uncharacterized protein LOC726990 isoform X2 | 0.3491918 | 0.7900076 | 0.1584704 | 0.3204690 | 0.0643546 |
| GB48685 | endothelin-converting enzyme-like 1-like isoform X1 | 0.3481051 | 0.3143372 | 0.0999660 | 0.8622472 | 0.0663038 |
| GB45358 | nuclear cap-binding protein subunit 2-like | 0.3468174 | 0.1324857 | 0.0623586 | -0.1019064 | -0.0363395 |
| GB49297 | protein eyes shut-like isoform X2 | 0.3338019 | -0.2293376 | -0.3155763 | 1.5346353 | 0.0553072 |
| GB51851 | diphthamide biosynthesis protein 7-like isoform X1 | 0.3151622 | -0.3917106 | -0.1136680 | -0.3304302 | -0.0659559 |
| GB48100 | forkhead box protein K2-like | 0.3078646 | 0.0502399 | -0.2868773 | 0.3823825 | -0.1059860 |
| GB53410 | nicotinamide riboside kinase | 0.3072171 | -0.0247020 | 0.0214844 | 1.5120289 | -0.0349848 |
| GB52361 | odorant receptor 2a | 0.3069290 | -2.2749522 | 0.3537685 | 0.6507751 | -0.4687087 |
| GB49771 | probable U3 small nucleolar RNA-associated protein 11-like isoform X1 | 0.3061378 | 0.0854558 | -0.0225643 | 3.6288168 | 0.0303111 |
| GB54350 | spermatogenesis-associated protein 6-like isoform X2 | 0.3017750 | -0.2294756 | 0.1341991 | 0.2227130 | 0.3339552 |
| GB52326 | chemosensory protein 4 precursor | 0.2774397 | 0.5027632 | 0.1834889 | 0.3130338 | 0.0521976 |
| 100576247 | frizzled-2-like, transcript variant X8 | 0.2713537 | 0.1589072 | -0.0487332 | -0.4928034 | 0.1490305 |
| GB41772 | heterochromatin protein 1-binding protein 3-like | 0.2688293 | -0.1073461 | -0.4365284 | 0.6386331 | -0.2152302 |
| GB46956 | homeobox protein B-H2-like | 0.2667862 | 0.0169211 | 0.0965548 | 0.9695841 | -0.3526284 |
| GB44120 | venom serine protease 34 isoform X2 | 0.2582506 | 0.7919654 | 0.2284436 | 0.2714301 | 0.1146043 |
| GB47515 | homeobox protein unplugged-like | 0.2533856 | -1.1508447 | 1.3223466 | 1.1131006 | 0.3932851 |
| GB52687 | GATA zinc finger domain-containing protein 4-like | 0.2478425 | 0.0848076 | -0.0351808 | 0.4253072 | -0.0704064 |
| GB54180 | segmentation protein paired | 0.2464985 | -0.3224775 | -0.0046032 | 0.3908382 | -0.0612394 |
| GB17991 | tyramine receptor | 0.2376049 | -0.4277596 | 0.1724761 | 0.1184470 | 0.3007527 |
| GB43643 | hepatic leukemia factor isoform X5 | 0.2300630 | -0.0885873 | -0.1806186 | 0.2619034 | -0.4952980 |
| GB45986 | scavenger receptor class B member 1 | 0.2281921 | 0.7366095 | 0.2183160 | 0.3183776 | 0.4549123 |
| 100578193 | uncharacterized protein LOC100578193 | 0.2265914 | -0.3755927 | 0.3792736 | 0.1916142 | -0.4536718 |
| 726803 | uncharacterized protein LOC726803 | 0.2215070 | 0.6745736 | -0.3232437 | -1.6983735 | 0.2396084 |
| GB52620 | paired box pox-meso protein isoform X1 | 0.2179205 | 0.3464557 | 0.5825832 | 0.4101937 | 0.0852373 |
| GB49410 | calmodulin-like | 0.2135726 | -0.0277908 | -0.0076846 | -1.4889160 | 0.3165611 |
| GB51295 | homeotic protein Sex combs reduced | 0.1994333 | -0.4054444 | 0.0757676 | 0.7076951 | -0.2371060 |
| GB49332 | brain-specific homeobox protein homolog | 0.1988772 | 0.7829426 | 0.0740177 | -0.0048292 | -0.3017339 |
| GB40967 | tyrosine hydroxylase | 0.1939191 | 0.2116776 | 0.0827130 | 0.0128425 | -0.1121341 |
| GB53374 | connectin isoform X2 | 0.1923983 | -0.4937854 | 0.0595257 | -0.3398432 | -0.6476856 |
| GB49973 | tachykinin-like peptides receptor 99D-like isoformX1 | 0.1903466 | 0.0684191 | -0.3541607 | 0.4937453 | -0.0304650 |
| GB42531 | zinc finger protein 470-like | 0.1840118 | -0.1266577 | 0.0929583 | 0.2507751 | 0.8348252 |
| GB52394 | odorant receptor 35 | 0.1836293 | 0.6743817 | -0.8881546 | 1.7333684 | 0.2586459 |
| GB40567 | serine protease nudel | 0.1791326 | -0.1805806 | 0.1324198 | -0.3540746 | -1.0144147 |
| GB47274 | UNC93-like protein-like | 0.1760126 | 0.0978842 | 0.0466488 | 0.2779259 | 0.0258282 |
| 100216325 | extra macrochaetae | 0.1721024 | 0.5375415 | -0.2121133 | 0.5896375 | 0.0534075 |
| GB40377 | uncharacterized protein LOC551717 | 0.1713617 | -0.4389868 | 0.3211403 | 0.5178133 | -0.5558815 |
| 100576903 | leucine-rich repeat-containing protein 15-like isoform 1 | 0.1676889 | -0.1640799 | -0.2865305 | 0.2111664 | -0.5339650 |
| GB50585 | alpha-2 adrenergic receptor-like | 0.1639323 | -1.3390923 | 0.1074478 | 0.4529004 | 0.3822319 |
| GB44017 | uncharacterized protein LOC411209 isoform X2 | 0.1625509 | 0.1474582 | 0.1569075 | 0.7036695 | 0.1786378 |
| GB40864 | titin-like | 0.1586610 | -0.2709110 | 0.1674224 | 0.3516364 | -0.5567276 |
| GB51441 | chromatin modification-related protein eaf-1-like isoform X1 | 0.1571053 | 0.1619194 | -0.0375848 | 0.2031531 | -0.3807868 |
| GB55297 | group XV phospholipase A2-like isoform X2 | 0.1568035 | -0.2536120 | 0.0416420 | 0.1940768 | 0.4299899 |
| GB42892 | uncharacterized protein LOC100578699 | 0.1562923 | -0.6872325 | 0.4138262 | -0.1097415 | 0.0715533 |
| 102654715 | general transcriptional corepressor trfA-like isoform X2 | 0.1559084 | 0.0078370 | 0.1479266 | 1.0733358 | -0.0399459 |
| GB55712 | oocyte zinc finger protein XlCOF6-like isoform 2 | 0.1480120 | 0.0952047 | 0.3215760 | -0.0374521 | 0.0247857 |
| GB50761 | chymotrypsin-1 | 0.1463405 | 1.0532771 | 0.3003517 | -0.1385155 | -0.3724792 |
| GB54775 | atrial natriuretic peptide-converting enzyme isoform X2 | 0.1460786 | 0.3743285 | -0.1566519 | -0.1456176 | -0.5334649 |
| 102655422 | uncharacterized protein LOC102655422 | 0.1385890 | -1.2954113 | 0.0707611 | 1.0059718 | -0.4904330 |
| GB54401 | elongation of very long chain fatty acids protein AAEL008004-like isoform X2 | 0.1336157 | 2.0899989 | -0.4185501 | 0.4709814 | 0.4028595 |
| GB51583 | kynurenine/alpha-aminoadipate aminotransferase, mitochondrial-like | 0.1294201 | -0.3657577 | -0.6117130 | 1.1668740 | -0.3777787 |
| 726761 | paired box protein Pax-2a-like isoform X2 | 0.1186476 | 0.0952047 | 0.1039544 | 0.5798076 | -0.3699653 |
| GB54748 | proteasome activator complex subunit 3-like | 0.1176236 | 0.7497801 | -0.0323915 | 0.7625122 | 0.0351167 |
| GB47018 | uncharacterized protein LOC724886 | 0.1157315 | -0.0164156 | -0.1022657 | 0.3806585 | -0.4249262 |
| 410557 | ATP synthase subunit d, mitochondrial | 0.1155345 | -0.2801539 | -0.1446580 | -0.1014718 | 0.1167322 |
| GB52755 | SET and MYND domain-containing protein 4-like isoform X1 | 0.1144706 | -0.4612234 | -0.0706732 | 0.1929210 | 0.2102913 |
| 551397 | 28S ribosomal protein S18a, mitochondrial isoform 2 | 0.1106191 | 0.0893239 | -0.0348236 | 5.0438999 | 0.0649919 |
| GB54292 | carbohydrate sulfotransferase 11-like | 0.1081089 | 0.6020144 | -0.1814898 | 0.0012975 | -0.7088836 |
| GB54802 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase-like isoform X4 | 0.1010411 | -0.0949371 | -0.0202266 | 0.2535593 | -0.1119863 |
| GB45977 | U11/U12 small nuclear ribonucleoprotein 25 kDa protein-like | 0.0973792 | -0.4071996 | 0.1883420 | 0.2946818 | -0.0274482 |
| GB44425 | coiled-coil domain-containing protein 104-like | 0.0950017 | 1.0012791 | -0.6552046 | 0.3222395 | -0.0112120 |
| GB42296 | peroxidase | 0.0944093 | 0.2603950 | 0.0552470 | 0.4341475 | -0.0883325 |
| GB49261 | uncharacterized protein LOC100576662 isoform X2 | 0.0939561 | 1.3148706 | -1.9023012 | 0.2238545 | 0.4933903 |
| GB40112 | uncharacterized protein LOC410462 | 0.0937457 | 0.1315488 | -0.1097024 | 1.2184136 | 0.0064863 |
| 102655945 | uncharacterized protein LOC102655945 | 0.0853910 | 1.0611690 | -0.0563931 | 0.0567426 | 0.4176833 |
| GB51846 | intraflagellar transport protein 46 homolog | 0.0853640 | 0.3026173 | 0.0688653 | 0.3690897 | -0.2026186 |
| GB41946 | cuticular protein analogous to peritrophins 3-D precursor | 0.0841994 | -0.4322829 | -0.1502015 | -0.1321769 | -0.3638915 |
| GB52186 | trypsin-1 | 0.0834781 | 0.3099989 | -0.0670718 | 0.9004645 | -0.5621062 |
| 725247 | probable inactive tRNA-specific adenosine deaminase-like protein 3-like | 0.0818763 | 0.0520746 | -0.0119089 | -0.1750042 | -0.0845647 |
| GB43690 | uncharacterized protein LOC727344 | 0.0773947 | 0.9254829 | 0.6988270 | -0.1254874 | 0.7329609 |
| GB51582 | aristaless-related homeobox protein isoform X1 | 0.0773385 | -0.7719506 | 0.1139744 | 0.0480020 | -0.5857722 |
| GB50650 | flocculation protein FLO11 isoform X3 | 0.0750604 | -0.4506516 | 0.3168894 | 0.2936290 | -0.3835473 |
| GB53126 | polypeptide N-acetylgalactosaminyltransferase 2-like isoform X1 | 0.0728786 | -0.1874685 | -0.0755599 | 0.0117675 | 0.2530391 |
| GB52465 | vitellogenin-2-like | 0.0706391 | -0.3641471 | -0.1829294 | 0.2532913 | 0.0637854 |
| 102656830 | uncharacterized protein LOC102656830 | 0.0694468 | -0.5795231 | 0.5620427 | -0.7711959 | 0.5290816 |
| GB55389 | spondin-1 isoform X1 | 0.0687016 | -0.1069032 | 0.0916424 | -0.2983582 | -0.0318121 |
| 100579019 | probable salivary secreted peptide-like | 0.0678622 | 1.3173295 | 0.3806096 | 0.9657867 | 0.7246295 |
| GB48935 | protein Star-like | 0.0656996 | 0.5328535 | 0.2258289 | -0.0290119 | 0.0975799 |
| GB45062 | protein apterous isoform X1 | 0.0635572 | -0.0040100 | 0.0667620 | 0.4329189 | -0.2189822 |
| GB51622 | heterogeneous nuclear ribonucleoprotein L isoform X1 | 0.0620070 | -0.3556979 | -0.0250075 | 0.0519768 | 0.1563677 |
| GB48039 | etoposide-induced protein 2.4-like isoform X2 | 0.0593876 | 0.1480499 | 0.0420642 | 0.3083053 | 0.0481981 |
| GB45218 | uncharacterized protein LOC408317 isoform 1 | 0.0590129 | -0.0701274 | 0.2031972 | 0.1401007 | 0.0130554 |
| GB55286 | homeobox protein SIX2 | 0.0564249 | 0.5427109 | 0.2187393 | -0.1002144 | 0.1124427 |
| GB41714 | uncharacterized protein LOC727150 isoform X2 | 0.0536204 | -0.3826573 | -0.2948150 | -0.6827824 | 0.1802347 |
| GB48943 | isocitrate dehydrogenase [NAD] subunit beta, mitochondrial-like | 0.0531532 | -0.2625487 | -0.3243812 | -1.0083422 | 0.1533136 |
| GB42736 | TM2 domain-containing protein CG10795-like | 0.0475663 | 0.5878244 | 0.0138815 | 0.3694662 | 0.3497229 |
| GB46213 | histone RNA hairpin-binding protein | 0.0454788 | 0.0631496 | 0.0987922 | 0.0075179 | 0.3001147 |
| GB42548 | protein kinase C-binding protein NELL1-like isoform X1 | 0.0434579 | 0.4743526 | 0.1943156 | 0.2260726 | -0.2723703 |
| GB46050 | protein king tubby-like | 0.0415573 | -0.0634324 | -0.0908351 | 0.0621844 | -0.0344164 |
| GB47259 | transcription initiation factor TFIID subunit 12 isoform X1 | 0.0382926 | 0.2916048 | 0.0603800 | 0.5179683 | 0.1507893 |
| GB50014 | coiled-coil domain-containing protein 111-like isoform X1 | 0.0375379 | -0.3719751 | 0.0627743 | -0.0525852 | 0.1564669 |
| 725329 | UPF0489 protein C5orf22 homolog isoform X3 | 0.0367872 | 0.3150846 | -0.0606647 | 0.0931977 | 0.1365816 |
| 725238 | histone H1B-like | 0.0309072 | 0.2753450 | 0.3068135 | 0.1248635 | -0.0022862 |
| GB46620 | uncharacterized protein C1orf112 homolog | 0.0276996 | 0.1393356 | 0.4834200 | 0.2695531 | 0.1281171 |
| GB43867 | uncharacterized protein PFB0765w-like | 0.0266379 | 0.4678793 | 0.3258482 | -0.0865333 | 0.0282067 |
| 724536 | uncharacterized protein LOC724536 | 0.0208600 | 0.3961154 | 0.1620852 | 0.9048472 | -0.2689525 |
| GB52644 | ATP synthase-coupling factor 6, mitochondrial | 0.0189552 | -0.1516881 | -0.2233460 | -0.0524388 | 0.2050755 |
| GB55014 | ras-related protein Rab-2-like | 0.0129896 | 0.3709669 | -0.0457940 | 0.5439276 | 0.0646793 |
| GB41419 | zinc finger protein 813-like | 0.0088166 | -0.4891554 | 0.0525762 | 0.3128437 | 0.0919048 |
Module 1
Table S26: List of all the genes in Module 1, ranked by their within-module connectivity, \(k\). The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
gene_list <- module_gene_list(1)
saveRDS(gene_list, file = "supplement/tab_S26.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB55823 | tRNA-splicing ligase RtcB homolog | 288.60578 | -0.0281965 | 0.1606359 | 0.0394358 | -0.0097862 |
| GB44900 | cullin-3 isoform X1 | 285.79810 | -0.0382861 | -0.0047554 | -0.0087679 | -0.0167811 |
| GB54956 | autophagy 1 | 283.90121 | 0.0479416 | -0.0109422 | -0.0038436 | 0.0293035 |
| GB43113 | serrate RNA effector molecule homolog isoform X1 | 274.93350 | -0.1828134 | 0.0298681 | 0.0456132 | 0.0070646 |
| GB45829 | vacuolar protein sorting-associated protein 4B isoformX1 | 272.20072 | 0.0626642 | 0.0262989 | 0.0405129 | 0.0741006 |
| GB47683 | LOW QUALITY PROTEIN: vacuolar protein sorting-associated protein 8 homolog | 271.82252 | -0.2813646 | 0.0640363 | 0.0200654 | 0.0620023 |
| GB41448 | rho GTPase-activating protein 26-like isoform X5 | 269.98626 | -0.1561438 | -0.0326420 | 0.0587879 | 0.0755708 |
| 725575 | axoneme-associated protein mst101(2)-like | 266.56485 | -0.2083017 | 0.0943316 | -0.0364053 | 0.0898112 |
| GB55512 | acidic fibroblast growth factor intracellular-binding protein isoform X1 | 264.66218 | -0.0103658 | 0.0249359 | 0.0105880 | -0.4332690 |
| GB43200 | tRNA (uracil-5-)-methyltransferase homolog A-like isoform X2 | 262.81057 | -0.1802649 | -0.0730565 | 0.0004482 | 0.0794535 |
| GB43952 | nitric oxide synthase-interacting protein homolog | 259.86949 | -0.1106611 | 0.1507171 | -0.2252511 | 0.0999624 |
| GB45823 | pre-mRNA-processing-splicing factor 8-like | 256.62337 | -0.0275978 | 0.0254559 | 0.0229017 | -0.0415693 |
| GB44359 | leucine-rich repeat-containing protein 16A-like isoform X4 | 251.88726 | -0.2150325 | 0.0201149 | 0.0425199 | 0.0791204 |
| GB53699 | survival of motor neuron-related-splicing factor 30-like isoform 2 | 250.81489 | -0.0532802 | 0.0106550 | -0.0371729 | 0.0489593 |
| GB44682 | catalase isoform 1 | 247.56249 | -0.2115596 | -0.0198514 | 0.0507368 | -0.0195449 |
| GB52979 | dentin sialophosphoprotein-like isoform X1 | 242.91001 | -0.1303534 | -0.0578291 | 0.0289106 | -0.0401034 |
| GB47599 | cytoplasmic dynein 1 light intermediate chain 1 isoform X2 | 242.47117 | -0.3757009 | 0.0127303 | 0.0578175 | 0.0349926 |
| GB44573 | ubiquitin carboxyl-terminal hydrolase 5-like | 241.46237 | 0.1447257 | 0.0672273 | -0.0857979 | 0.0598318 |
| GB50370 | histone-lysine N-methyltransferase SETD1B-like isoform 1 | 240.88326 | -0.1331707 | 0.1015549 | 0.6914511 | 0.0565033 |
| GB41911 | procollagen-lysine,2-oxoglutarate 5-dioxygenase 3-like isoform X1 | 240.26407 | -0.1112528 | 0.0986711 | 0.0288154 | 0.4948914 |
| GB44594 | tyrosine-protein kinase hopscotch isoform X2 | 240.01837 | -0.1512548 | 0.0282488 | -0.0757608 | 0.0199088 |
| GB49111 | neuropathy target esterase sws isoform X3 | 239.64807 | -0.0757516 | -0.0047253 | -0.0095098 | 0.2978976 |
| GB52662 | probable actin-related protein 2/3 complex subunit 2 isoform 2 | 237.85199 | -0.0906784 | 0.0390845 | 0.0221270 | 0.0512174 |
| GB48925 | superkiller viralicidic activity 2-like 2-like isoform X2 | 234.51059 | -0.1250507 | 0.0268830 | 0.0365259 | -0.0074083 |
| GB50757 | KH domain-containing, RNA-binding, signal transduction-associated protein 3-like isoformX2 | 232.04054 | -0.1457952 | -0.0727868 | 0.0413845 | 0.0402818 |
| 724928 | tubulin-specific chaperone C-like isoform 1 | 232.01258 | -0.0979867 | 0.0556192 | -0.0244316 | 0.1072274 |
| GB56004 | ATPase family AAA domain-containing protein 1-A-like | 229.97172 | 0.0491721 | 0.0287752 | 0.0581703 | 0.0437436 |
| GB55941 | hemK methyltransferase family member 1-like isoform X2 | 229.96678 | -0.0639887 | 0.1344013 | -0.0585299 | 0.0227723 |
| GB53725 | splicing factor 3B subunit 1-like isoform X2 | 227.26120 | -0.3550024 | -0.0844896 | 0.0817055 | -0.0164484 |
| GB49918 | optineurin isoform X2 | 226.53292 | -0.3600699 | 0.0370622 | 0.0476319 | 0.0622934 |
| GB42141 | probable medium-chain specific acyl-CoA dehydrogenase, mitochondrial-like | 225.66044 | 0.6614338 | -0.1489981 | -0.0676639 | 0.1896882 |
| GB49425 | ATP-dependent zinc metalloprotease YME1 homolog isoform X3 | 225.13635 | -0.4503013 | 0.1263263 | 0.0349089 | 0.0999659 |
| GB44423 | vacuolar fusion protein MON1 homolog A-like | 224.89964 | -0.0033257 | -0.0179326 | 0.0422244 | 0.0122336 |
| GB51133 | uncharacterized protein LOC725950 isoform X7 | 224.34524 | -0.2949259 | -0.0250259 | 0.2953370 | -0.0581774 |
| GB49943 | eukaryotic translation initiation factor 3 subunit D | 223.69559 | 0.1541782 | 0.0548449 | 0.0315715 | -0.0248784 |
| GB43707 | smallminded | 223.35421 | -0.5113509 | 0.0899614 | 0.0309637 | 0.0359696 |
| GB43172 | cyclin-dependent kinase 11B isoform X1 | 222.96837 | -0.0206602 | -0.0559101 | -0.0643595 | -0.0510414 |
| GB47089 | BRCA1-A complex subunit Abraxas-like isoform X2 | 222.94741 | -0.0656233 | 0.2064747 | 0.0365300 | 0.0739280 |
| GB47208 | mitogen-activated protein kinase kinase kinase 10 isoform X4 | 221.51290 | -0.1552847 | 0.0183887 | 0.0324778 | -0.0443445 |
| GB48347 | WASH complex subunit strumpellin-like isoform X3 | 220.70903 | -0.3686723 | 0.1050430 | -0.0075903 | 0.0235721 |
| GB44556 | uncharacterized protein LOC411962 isoform X2 | 220.16678 | -0.1133288 | 0.1546915 | 0.0027307 | 0.0339605 |
| GB50854 | AP-1 complex subunit beta-1 | 219.38155 | -0.1349754 | -0.0211709 | -0.0270254 | -0.0266302 |
| GB55540 | zinc finger MYM-type protein 3-like | 219.30291 | -0.0118923 | -0.0201705 | 0.0769080 | 0.1084151 |
| GB46636 | probable exonuclease mut-7 homolog isoform X2 | 219.01449 | -0.2838153 | -0.1186433 | 0.0074181 | 0.0254409 |
| GB40507 | nucleolar protein 10 | 218.40981 | -0.2600232 | 0.1445714 | -0.0551367 | 0.1223630 |
| GB54228 | SUMO-activating enzyme subunit 2 isoform X1 | 217.33099 | 0.0566474 | 0.1842158 | 0.1083653 | -0.0153830 |
| GB54389 | copper-transporting ATPase 1 isoform X2 | 217.11573 | 0.1360450 | -0.0242164 | 0.0349226 | -0.0757144 |
| GB44516 | exostasin 2, transcript variant X2 | 217.07220 | -0.1341388 | -0.1048453 | 0.0576125 | 0.0588822 |
| GB41488 | ankyrin repeat and LEM domain-containing protein 2-like isoform X4 | 216.31840 | -0.3975618 | 0.0422519 | 0.0358214 | 0.1054610 |
| GB50042 | vesicular integral-membrane protein VIP36 | 215.91764 | 0.2931243 | -0.0695297 | 0.5128282 | 0.0768173 |
| GB47110 | methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial-like | 215.49170 | -0.0366004 | -0.0681734 | -0.0549869 | -0.7324760 |
| GB49428 | tuberin isoform X2 | 215.21096 | -0.1966563 | -0.0427339 | 0.0649788 | 0.0837811 |
| GB51360 | integrator complex subunit 1-like, transcript variant X2 | 214.63924 | -0.0009533 | -0.1018687 | -0.0341014 | 0.0959737 |
| GB45873 | ubiquitin conjugation factor E4 A-like isoform X2 | 214.47969 | -0.1298061 | -0.0670541 | -0.0187154 | 0.0912066 |
| GB41024 | U3 small nucleolar RNA-associated protein 6 homolog isoform X2 | 214.21983 | -0.0754706 | 0.2934448 | -0.0471835 | -0.0133720 |
| GB40711 | RING finger protein 10-like | 214.18339 | -0.2255345 | -0.0612001 | 0.0301634 | 0.0202273 |
| GB50872 | general transcription factor IIF subunit 2 isoformX1 | 213.92042 | -0.1533262 | 0.1310806 | 0.0659204 | 0.0550357 |
| GB42977 | DNA polymerase delta catalytic subunit isoform X2 | 212.90650 | -0.1488819 | 0.0865535 | -0.0887230 | 0.0205718 |
| GB54054 | LOW QUALITY PROTEIN: ubiquitin carboxyl-terminal hydrolase 7 | 210.77828 | 0.0951204 | 0.0190603 | 0.0652836 | -0.2480764 |
| GB44421 | replication protein A 70 kDa DNA-binding subunit isoform X2 | 210.01165 | -0.2826954 | 0.0303812 | -0.0334162 | 0.1465698 |
| GB46511 | protein bric-a-brac 2 isoform X1 | 209.84256 | -0.2726323 | 0.2269248 | 0.1011703 | 0.1941534 |
| GB46065 | Hermansky-Pudlak syndrome 5 protein homolog isoform X1 | 208.51637 | -0.3335396 | -0.0692699 | -0.0184378 | -0.0121288 |
| GB49749 | uncharacterized protein C12orf4 homolog | 208.43198 | 0.1077336 | 0.1128373 | 0.0514064 | 0.0328803 |
| GB42159 | uncharacterized exonuclease C637.09-like isoform X3 | 208.00661 | -0.2291293 | 0.1394666 | 0.0405295 | 0.0173193 |
| GB46431 | eukaryotic translation initiation factor 3 subunit A | 207.75211 | -0.0484745 | 0.0738027 | 0.0359871 | -0.0801569 |
| GB42058 | helicase SKI2W | 207.73875 | -0.3389229 | 0.0170163 | 0.0148386 | -0.0011292 |
| GB55476 | oxysterol-binding protein-related protein 9-like isoform X4 | 207.60126 | -0.1213446 | 0.0759970 | 0.0816787 | -0.2342863 |
| GB49296 | zinc finger CCCH domain-containing protein 13-like | 207.29654 | -0.3953375 | 0.0585999 | 0.0106809 | -0.0088986 |
| GB42448 | mediator of RNA polymerase II transcription subunit 15-like isoform X2 | 206.68794 | -0.4547626 | 0.0176299 | 0.2826344 | 0.1883594 |
| 410869 | vacuolar-sorting protein SNF8 isoform X1 | 206.50165 | -0.1110148 | 0.0430158 | 0.0852174 | 0.1127796 |
| GB50847 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-like | 206.23119 | -0.0544846 | 0.1186289 | 0.0439726 | 0.0072304 |
| GB50287 | RAD50-interacting protein 1-like isoform X1 | 205.39572 | -0.2573657 | -0.0043303 | 0.1784503 | -0.0248772 |
| GB50276 | dual specificity mitogen-activated protein kinase kinase 4 | 205.10168 | 0.1950044 | 0.0493514 | 0.0517128 | 0.0333314 |
| GB54194 | charged multivesicular body protein 4b isoform X1 | 205.02779 | 0.0124217 | 0.0190265 | -0.0035026 | 0.0848020 |
| GB45008 | angio-associated migratory cell protein-like isoform X2 | 204.58184 | -0.0693296 | 0.0848315 | -0.1822455 | 0.0321644 |
| GB55459 | general transcription factor IIH subunit 1 isoform X1 | 203.92803 | -0.0564831 | 0.1354081 | 0.0792959 | 0.0339948 |
| GB45683 | WD repeat-containing protein 3-like | 203.88116 | -0.2248092 | 0.0414917 | 0.0189259 | 0.0605902 |
| GB41025 | chromobox protein homolog 5-like | 203.64027 | -0.3577102 | -0.0611846 | 0.0556368 | -0.2955834 |
| GB51246 | mediator of RNA polymerase II transcription subunit 23 isoform X2 | 203.42594 | -0.0531331 | -0.0438789 | 0.0418241 | -0.0064390 |
| GB54087 | zinc finger FYVE domain-containing protein 19-like | 203.27090 | -0.1623556 | -0.0450021 | 0.0891825 | 0.0966665 |
| GB54425 | cullin-associated NEDD8-dissociated protein 1-like isoform X2 | 202.73240 | 0.2014709 | 0.0849647 | -0.0221812 | 0.0332585 |
| GB41258 | BTB/POZ domain-containing adapter for CUL3-mediated RhoA degradation protein 3-like | 202.63989 | -0.1244124 | 0.0601458 | -0.0383048 | 0.0104664 |
| GB51495 | u4/U6.U5 tri-snRNP-associated protein 1-like, transcript variant X2 | 202.47328 | -0.2072260 | 0.0624135 | 0.0445140 | 0.1140704 |
| GB47575 | tyrosine-protein phosphatase corkscrew isoform X4 | 202.14336 | -0.0520171 | -0.0686207 | 0.0453436 | 0.0411802 |
| GB41293 | histone acetyltransferase KAT8 isoform X1 | 201.80732 | 0.0731326 | -0.0280351 | 0.4838555 | 0.0243201 |
| GB54589 | YTH domain family protein 3-like isoform X1 | 201.61658 | -0.2767910 | 0.0481742 | 0.1163326 | 0.0103777 |
| GB45114 | HEAT repeat-containing protein 1 isoform X2 | 201.61325 | -0.0382402 | 0.0857539 | -0.3060003 | 0.0492955 |
| GB44690 | serine/threonine-protein phosphatase 4 regulatory subunit 3 isoform X2 | 200.87975 | -0.0678411 | 0.2092908 | 0.0480516 | 0.1631517 |
| GB50409 | splicing factor 3A subunit 1 isoformX1 | 200.84030 | -0.2564942 | 0.1671163 | -0.0032251 | -0.0553593 |
| GB43192 | transcription initiation factor TFIID subunit 6 isoform X2 | 200.83407 | -0.0035335 | -0.0810491 | -0.0059793 | -0.0502864 |
| GB53829 | CD2 antigen cytoplasmic tail-binding protein 2 homolog | 200.78864 | -0.2900452 | 0.0257855 | -0.0384880 | -0.0952961 |
| GB44912 | lethal(3)malignant brain tumor-like protein 3-like isoform X6 | 200.71246 | -0.3218606 | 0.0963412 | 0.0702666 | 0.1880399 |
| GB47102 | general vesicular transport factor p115 | 200.22598 | -0.2489558 | 0.0260273 | -0.0391379 | 0.3523676 |
| GB52559 | LMBR1 domain-containing protein 2 homolog | 199.81537 | 0.0894252 | -0.0682850 | 0.0421673 | 0.0129771 |
| GB43122 | probable RNA-binding protein 19-like isoform X2 | 199.50308 | 0.1037877 | 0.0254709 | 0.0230659 | 0.0584319 |
| GB46972 | vacuolar protein sorting-associated protein 52 homolog isoform 1 | 199.01673 | -0.3521252 | -0.0413197 | -0.0537183 | -0.0007593 |
| GB40578 | sodium/hydrogen exchanger 7 isoform X1 | 197.99334 | 0.1602845 | -0.0597625 | 0.1297220 | -0.0419840 |
| GB55413 | zinc transporter 9-like | 197.33188 | -0.1174266 | 0.0917137 | 0.0326560 | 0.0912740 |
| GB51717 | WASH complex subunit 7-like | 196.82193 | 0.0976007 | 0.0871875 | 0.0488481 | -0.1357646 |
| GB51954 | sec1 family domain-containing protein 2-like isoform X2 | 196.77792 | -0.1159123 | -0.0372640 | 0.0298109 | 0.0810476 |
| GB47680 | TATA-binding protein-associated factor 172-like isoform X2 | 196.59706 | -0.2510289 | 0.0557936 | 0.4483637 | -0.2930443 |
| GB52641 | glyoxalase domain-containing protein 4-like | 196.56385 | -0.4062763 | 0.0783375 | 0.0006743 | 0.0573930 |
| GB48101 | importin-4-like | 196.49388 | 0.3244796 | 0.1313727 | 0.6063033 | 0.0647293 |
| GB49047 | 26S protease regulatory subunit 6A | 196.44539 | 0.1870912 | -0.0769411 | -0.0688529 | 0.0124138 |
| GB54045 | ras-related protein Rab-40C-like isoform 2 | 196.44107 | -0.0714702 | -0.0392858 | -0.0236941 | 0.0779570 |
| GB40893 | trafficking protein particle complex subunit 8-like isoform X3 | 196.40522 | -0.4376881 | 0.0147667 | -0.0150959 | -0.2189324 |
| 100578159 | zinc finger matrin-type protein CG9776-like isoform X4 | 196.30337 | -0.2787936 | -0.0067094 | -0.1794822 | 0.6734605 |
| GB47093 | activator of basal transcription 1-like | 196.00855 | -0.2592536 | 0.0584833 | -0.0210939 | 0.0941891 |
| GB44486 | tyrosine-protein phosphatase non-receptor type 21 isoform X2 | 195.67834 | -0.0424408 | -0.0279357 | 0.0603428 | 0.0516163 |
| GB49468 | RRP12-like protein-like isoform X2 | 195.37538 | -0.3181060 | 0.0535266 | 0.0751676 | -0.0371262 |
| GB43565 | OTU domain-containing protein 5-A-like | 195.02494 | 0.1160859 | 0.1349821 | 0.0564931 | 0.0420354 |
| GB53614 | RNA-binding protein 26 isoform X2 | 195.00729 | -0.0370210 | 0.0914187 | 0.0423985 | 0.0450740 |
| GB54328 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase-like | 194.81477 | -0.2904883 | -0.0153050 | 0.0555989 | 0.0358194 |
| GB41239 | trichohyalin | 194.80269 | -0.4969900 | 0.1078321 | 0.0416462 | -0.0650865 |
| GB41795 | vacuolar protein sorting-associated protein 33B isoform X2 | 194.64502 | 0.0184606 | -0.0756184 | -0.0092399 | 0.0720473 |
| GB54526 | mitotic spindle assembly checkpoint protein MAD1 | 194.42503 | -0.0968978 | 0.1590043 | 0.0563812 | 0.1427241 |
| GB45184 | 2-hydroxyacyl-CoA lyase 1-like | 194.01437 | 0.2399532 | -0.1341564 | -0.3798903 | -0.0909855 |
| GB40361 | sister chromatid cohesion protein PDS5 homolog B-B-like isoform X1 | 193.12195 | -0.2778751 | 0.0418388 | -0.0040056 | -0.0692047 |
| GB54995 | Rho-associated, coiled-coil containing protein kinase 2, transcript variant X2 | 193.00300 | -0.2527626 | -0.0531265 | 0.0774855 | -0.0314964 |
| GB50952 | E3 ubiquitin-protein ligase RNF13-like isoform X2 | 192.53325 | -0.0427018 | -0.0506680 | 0.0632524 | 0.1139598 |
| GB10936 | U4/U6 small nuclear ribonucleoprotein Prp3 | 192.47652 | -0.0832154 | 0.1059277 | 0.0392885 | 0.0493244 |
| GB47660 | alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase B-like isoform X3 | 192.16032 | -0.0683519 | 0.0627941 | 0.0415849 | 0.1806669 |
| GB40880 | DDB1- and CUL4-associated factor-like 1-like isoform X1 | 191.99601 | -0.1845890 | 0.0274321 | 0.0870227 | 0.0344227 |
| GB49654 | zinc finger protein 830-like | 191.79217 | -0.5377447 | 0.0510624 | 0.0420019 | 0.1165216 |
| GB45214 | zinc finger FYVE domain-containing protein 16 isoform X5 | 191.54102 | -0.2169496 | 0.0410720 | 0.0791372 | 0.1653875 |
| GB49409 | tetratricopeptide repeat protein 7B-like, transcript variant X5 | 191.37636 | -0.2374575 | -0.0092338 | 0.0054186 | 0.0171132 |
| GB43540 | uncharacterized protein LOC552071 | 191.18104 | -0.3225314 | 0.0399793 | 0.0517002 | 0.0871493 |
| GB47904 | 26S protease regulatory subunit 7 | 191.16782 | 0.1931016 | 0.0261522 | -0.0884919 | -0.0216421 |
| GB43124 | uncharacterized protein C19orf47 homolog | 190.69983 | -0.1270242 | 0.0090097 | -0.1110097 | 0.1137965 |
| GB47888 | bifunctional protein NCOAT-like isoform X2 | 190.61237 | -0.1794708 | 0.0116590 | -0.1209920 | 0.0551990 |
| GB55056 | spermatogenesis-associated protein 20 isoform X2 | 189.60767 | -0.4202731 | -0.1339388 | 0.2933127 | 0.0762342 |
| GB55644 | uncharacterized LOC409221, transcript variant X3 | 189.46212 | 0.0184559 | 0.0382613 | 0.0226602 | 0.0069678 |
| 410886 | E3 ubiquitin-protein ligase UBR2-like | 189.45383 | -0.0657603 | -0.1132438 | 0.1327208 | 0.0552951 |
| GB54718 | DNA repair protein REV1 | 188.92044 | -0.1590842 | 0.0141426 | 0.1022615 | 0.1018506 |
| GB46344 | ubiquitin carboxyl-terminal hydrolase 8-like isoform X2 | 188.90772 | 0.1338219 | 0.0127657 | 0.0673908 | 0.2682462 |
| GB49490 | actin-like protein 87C-like | 188.71480 | 0.2739357 | 0.0014613 | 0.0567098 | 0.1218011 |
| GB40741 | PAX-interacting protein 1-like | 187.83922 | -0.1494487 | 0.0936160 | -0.1602269 | 0.0859703 |
| GB45436 | coiled-coil and C2 domain-containing protein 1-like isoform X2 | 187.64562 | -0.2334624 | -0.0042491 | -0.3146303 | 0.1725393 |
| GB40948 | uncharacterized protein LOC412397 isoform X2 | 187.60150 | -0.1195504 | -0.1987567 | 0.0329359 | -0.0120353 |
| GB42841 | very long-chain specific acyl-CoA dehydrogenase, mitochondrial-like | 187.44986 | 0.1025917 | -0.0557134 | 0.1898248 | -0.2027086 |
| 726952 | ER degradation-enhancing alpha-mannosidase-like 1 | 187.41257 | -0.2617900 | 0.0639915 | -0.0160886 | -0.0295248 |
| GB46070 | protein SMG5-like | 187.27829 | -0.1229102 | -0.0925536 | 0.3059350 | -0.0315365 |
| GB45452 | AP-3 complex subunit mu-1-like isoform X1 | 187.04835 | 0.0655556 | 0.0255746 | -0.0314282 | -0.0108713 |
| GB50607 | grpE protein homolog 1, mitochondrial | 187.03757 | 0.0096856 | -0.0449069 | -0.0250354 | 0.0850403 |
| GB43637 | protein asunder homolog | 186.75665 | 0.2475874 | 0.1792277 | -0.0441214 | -0.0418747 |
| GB50231 | protein RMD5 homolog A-like | 186.62899 | 0.3061106 | -0.0209362 | 0.0333957 | 0.0617826 |
| GB40744 | squamous cell carcinoma antigen recognized by T-cells 3 | 186.59024 | 0.0031934 | 0.1430022 | 0.0765439 | 0.0040247 |
| GB50945 | dentin sialophosphoprotein-like | 186.47581 | -0.2403981 | 0.2168386 | -0.0885244 | -0.0121863 |
| GB50067 | charged multivesicular body protein 3 | 186.29749 | 0.0373942 | -0.0392116 | -0.0473283 | 0.0914076 |
| GB51588 | actin-related protein 2-like isoform X5 | 186.02492 | -0.1207530 | -0.0135926 | 0.0840078 | 0.0924636 |
| GB47440 | dynamin related protein 1 | 185.71493 | 0.3390192 | 0.1217222 | 0.0267123 | 0.0409860 |
| GB42750 | LOW QUALITY PROTEIN: PHD finger protein 14-like | 185.70490 | -0.0904365 | 0.1562576 | 0.0224898 | 0.1213014 |
| GB42223 | elongation factor G, mitochondrial-like | 185.66155 | -0.2882916 | 0.0108843 | -0.0950271 | 0.7205432 |
| GB42739 | xanthine dehydrogenase isoform X4 | 185.64800 | -0.1405292 | -0.1027501 | -0.0623907 | 0.0629294 |
| GB48532 | protein zer-1 homolog isoform X2 | 185.28231 | -0.0308770 | 0.0162875 | 0.1010409 | 0.0689667 |
| GB55550 | ras GTPase-activating protein 1-like, transcript variant X3 | 185.04993 | 0.0439635 | 0.1644054 | 0.0821610 | 0.0309474 |
| GB48631 | probable complex I intermediate-associated protein 30, mitochondrial-like | 184.95905 | 0.0568233 | 0.0176154 | 0.0873688 | 0.0771332 |
| GB50083 | pre-mRNA-splicing factor SPF27 isoform X1 | 184.87477 | 0.0208596 | 0.0744052 | 0.0270616 | 0.0235241 |
| GB47333 | probable glutamate–tRNA ligase, mitochondrial-like | 184.87031 | -0.3534262 | 0.0248370 | -0.0073916 | 0.2747849 |
| GB46028 | E3 ubiquitin-protein ligase RNF14-like | 184.64677 | -0.4690063 | 0.0376000 | 1.1864935 | 0.2093782 |
| GB43178 | WD repeat-containing protein mio-B isoform X2 | 184.57653 | 0.0293342 | 0.0525459 | 0.0684298 | 0.0325748 |
| GB41080 | ubiquitin carboxyl-terminal hydrolase 14-like isoform 2 | 184.34094 | 0.1225644 | -0.0941514 | 0.0362429 | -0.0218409 |
| GB45113 | mitochondrial inner membrane protein OXA1L-like | 182.83456 | -0.3997153 | -0.0529569 | -0.0615654 | -0.0646304 |
| GB46026 | mitochondrial import receptor subunit TOM70 | 182.74842 | 0.1008186 | 0.0267759 | -0.1440612 | 0.0690353 |
| GB42452 | mediator of RNA polymerase II transcription subunit 27 | 182.52298 | -0.2732879 | 0.1047517 | 0.1002314 | 0.1012910 |
| GB51699 | WD repeat-containing protein 43-like | 182.24772 | 0.2668500 | 0.1496757 | 0.1169627 | 0.0528346 |
| GB45739 | 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial-like isoform 1 | 182.21100 | 0.1174785 | 0.0235244 | -0.0381293 | -0.1973933 |
| GB47420 | nuclear RNA export factor 1-like isoform 2 | 182.07859 | -0.3060885 | 0.0469920 | 0.0715271 | 0.1336460 |
| GB46118 | cerebellar degeneration-related protein 2-like isoform X1 | 181.50554 | -0.1810810 | 0.1412101 | 0.1458258 | -0.0011423 |
| GB40466 | ATP-binding cassette sub-family F member 2-like isoform X1 | 181.49253 | -0.2929678 | 0.1022491 | 0.0135726 | 0.0676464 |
| GB45345 | zinc finger MYND domain-containing protein 11-like isoform X2 | 181.47460 | -0.2657453 | -0.0096304 | 0.0320374 | -0.0016745 |
| GB54780 | protein brunelleschi-like isoform X2 | 181.36263 | -0.0920858 | 0.0445131 | 0.0093270 | -0.0018704 |
| GB52883 | FAM203 family protein GA19338-like | 180.65742 | -0.3794956 | -0.0715496 | 0.0201639 | 0.0823561 |
| GB52978 | TBC1 domain family member 15 | 180.57903 | -0.0698828 | -0.0207859 | 0.0459486 | 0.1170165 |
| GB48457 | peptidylprolyl isomerase domain and WD repeat-containing protein 1 isoform X3 | 180.39663 | -0.2662506 | 0.0330535 | 0.1503931 | 0.0980952 |
| GB44048 | endoplasmic reticulum lectin 1-like isoform X4 | 179.73712 | 0.2131686 | 0.0341315 | -0.0493331 | 0.0383247 |
| GB43448 | calpain-B | 179.65040 | -0.1504593 | -0.1893015 | 0.0950542 | -0.0553624 |
| GB49027 | sulfide:quinone oxidoreductase, mitochondrial-like isoform X2 | 179.61486 | -0.2412945 | 0.0622571 | 0.0283813 | 0.0150266 |
| GB41713 | TATA element modulatory factor-like isoform X2 | 179.52439 | -0.5942016 | -0.1334045 | 0.0817136 | 0.0338489 |
| GB52536 | LOW QUALITY PROTEIN: proteasome-associated protein ECM29 homolog | 179.51337 | -0.3709278 | -0.1049523 | 0.0249237 | 0.0360125 |
| GB47250 | THO complex subunit 1-like isoform X2 | 179.50239 | -0.4606453 | -0.0355974 | -0.0108644 | 0.1065647 |
| GB46034 | syntaxin-12 | 179.27246 | -0.5643866 | 0.1050736 | 0.0595323 | -0.0092484 |
| GB46121 | ubiquitin fusion degradation protein 1 homolog isoform X2 | 179.20842 | 0.1628130 | -0.0690992 | -0.0597555 | 0.0616813 |
| GB45831 | beta-parvin-like | 179.06186 | 0.0981391 | -0.0717356 | 0.0405107 | 0.0649034 |
| GB55931 | double-strand-break repair protein rad21 homolog isoform X1 | 178.51700 | 0.0934565 | -0.0059451 | 0.1276780 | 0.0174615 |
| GB51496 | uncharacterized protein C17orf85 homolog | 178.48117 | -0.4429990 | 0.0208189 | 0.0856665 | 0.0212191 |
| GB52628 | heat shock factor protein isoform X3 | 178.38518 | -0.4029739 | -0.1067077 | 0.1281488 | -0.0121320 |
| GB46594 | striatin-interacting proteins 2-like isoform X1 | 178.31577 | -0.0522705 | -0.0782125 | -0.0809841 | 0.1447182 |
| GB46074 | survival motor neuron protein-like | 178.23320 | 0.0318356 | -0.1049528 | -0.1251078 | -0.0021880 |
| GB47329 | WD and tetratricopeptide repeats protein 1-like isoform X2 | 178.16797 | -0.1775274 | -0.0162210 | 0.0006628 | 0.1730579 |
| GB42204 | rho guanine nucleotide exchange factor 3-like | 177.95959 | -0.3994772 | -0.0160166 | -0.0599380 | 0.0209071 |
| GB45868 | F-box/WD repeat-containing protein 7 isoform X2 | 177.79490 | -0.2331125 | 0.0752654 | 0.0799164 | -0.1496531 |
| GB48544 | coiled-coil domain-containing protein 43-like | 177.54881 | -0.0017113 | 0.0318609 | 0.2139969 | 0.0935069 |
| GB50345 | probable phosphorylase b kinase regulatory subunit alpha-like isoform X4 | 177.04077 | -0.1970543 | -0.1368905 | 0.1315939 | -0.0200568 |
| GB49040 | KAT8 regulatory NSL complex subunit 2 isoform X5 | 176.85922 | 0.0628656 | 0.0015017 | 0.0950085 | 0.1013152 |
| GB50587 | uncharacterized protein LOC410622 | 176.82342 | -0.1294509 | -0.0494395 | 0.0295796 | 0.0206181 |
| GB43464 | lysosomal Pro-X carboxypeptidase-like | 176.81252 | -0.0321209 | -0.1409916 | -0.0328756 | -0.3729907 |
| GB49651 | exocyst complex component 3 | 176.78523 | 0.3444830 | 0.0335798 | 0.0782172 | 0.0560180 |
| GB47161 | pre-mRNA-splicing factor 18-like | 176.68527 | -0.3164830 | -0.0414798 | 0.0783095 | 0.0497773 |
| GB53617 | sorting and assembly machinery component 50 homolog | 176.58814 | 0.2471963 | 0.0396593 | 0.0600269 | -0.0000683 |
| GB44439 | eukaryotic initiation factor 4A-III-like isoform 1 | 176.36635 | 0.2454471 | 0.0368400 | 0.0224123 | 0.1284649 |
| GB55333 | N-alpha-acetyltransferase 35, NatC auxiliary subunit isoform X2 | 176.32691 | -0.2174214 | -0.0214850 | 0.1622942 | 0.0288509 |
| GB46061 | RNA-binding protein 28-like | 176.30769 | -0.5296071 | 0.0384121 | -0.0595057 | 0.1182992 |
| GB41244 | inositol polyphosphate 5-phosphatase K-like isoformX1 | 176.00550 | 0.4250308 | -0.1333282 | -0.3800855 | 0.0761223 |
| GB52848 | titin-like | 175.91179 | -0.7125301 | -0.0077780 | 0.8302290 | 0.3280266 |
| GB46036 | periodic tryptophan protein 2 homolog | 175.76898 | -0.5643656 | 0.1320948 | -0.0433318 | 0.0847922 |
| GB50145 | ubiquitin domain-containing protein 2-like | 175.67101 | -0.0961307 | -0.0170959 | -0.0665807 | 0.0997034 |
| GB46927 | F-box only protein 21-like | 175.66482 | -0.5308469 | -0.0239916 | 0.6896461 | 0.8500469 |
| GB51464 | rho GTPase-activating protein 44-like isoform X2 | 175.50768 | -0.0356736 | 0.0748345 | -0.0162261 | 0.1120328 |
| GB44172 | protein CASC3-like isoform X2 | 175.36510 | -0.2780037 | 0.0571639 | 0.1046004 | 0.0705342 |
| GB46655 | splicing factor U2AF 50 kDa subunit isoform X1 | 175.34594 | 0.2411086 | 0.0628230 | -0.0157794 | -0.0181715 |
| GB49596 | NEDD8-activating enzyme E1 catalytic subunit-like | 175.33079 | 0.0017760 | 0.0725714 | -0.0368334 | 0.0216606 |
| GB49974 | 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial-like isoform X1 | 175.25673 | -0.3626690 | 0.0262583 | 0.0964368 | -0.0016298 |
| GB46211 | zinc finger protein 665-like isoform X3 | 174.89780 | -0.0371873 | 0.0919620 | 0.0388458 | 0.0879322 |
| GB42525 | zinc finger protein ZPR1 | 174.77262 | -0.1756571 | 0.0799344 | -0.1391161 | 0.0889572 |
| GB49423 | probable queuine tRNA-ribosyltransferase | 174.76994 | -0.0692681 | -0.0473035 | 0.0835788 | 0.1350037 |
| GB51806 | coiled-coil domain-containing protein 93 isoform X2 | 174.66102 | -0.0740182 | -0.0777750 | -0.0645862 | 0.2814372 |
| GB45534 | exocyst complex component 7 | 174.53202 | -0.1691080 | 0.0860970 | 0.1086326 | 0.0380245 |
| GB47192 | exosome complex component MTR3-like isoform X2 | 174.45096 | 0.0893465 | 0.0284133 | 0.0095640 | 0.1473212 |
| GB54583 | H/ACA ribonucleoprotein complex non-core subunit NAF1-like | 174.21282 | -0.5948602 | 0.0305381 | -0.0724988 | -0.0478107 |
| GB40858 | probable ATP-dependent RNA helicase DDX47-like isoform 1 | 174.21195 | -0.2341246 | 0.0555878 | -1.0745571 | 0.0215981 |
| GB44010 | zinc finger protein 23-like isoform 1 | 174.09862 | -0.4749669 | 0.2433766 | -0.4748139 | 0.0548570 |
| GB49244 | charged multivesicular body protein 5-like | 174.09745 | 0.2531362 | 0.0673163 | 0.0949334 | 0.0353168 |
| GB46494 | Golgi reassembly-stacking protein 2-like | 174.00731 | 0.1043000 | 0.0783469 | 0.5963635 | -0.0720191 |
| GB40267 | probable 39S ribosomal protein L45, mitochondrial | 173.61348 | -0.3108193 | 0.0808213 | 0.1174777 | 0.0445115 |
| GB41968 | protein max isoform 2 | 173.47923 | 0.1398838 | -0.0192584 | 0.1033002 | 0.0411996 |
| GB48170 | ran GTPase-activating protein 1-like isoform X1 | 173.43099 | -0.1000298 | 0.1880882 | -0.0203595 | 0.1301734 |
| GB52673 | gamma-tubulin complex component 6-like isoform X1 | 173.25955 | -0.3170258 | 0.1045833 | 0.0360074 | 0.1465033 |
| GB49776 | retinoblastoma-binding protein 5-like isoformX1 | 173.22991 | -0.1295567 | 0.0659218 | -0.0421685 | 0.0732333 |
| GB41902 | pre-rRNA-processing protein TSR1 homolog | 173.17493 | -0.2497342 | 0.1009075 | 0.0289703 | 0.0035600 |
| GB40559 | CDK5 regulatory subunit-associated protein 3-like | 173.01764 | -0.1834198 | 0.0129343 | -0.0847038 | -0.0949503 |
| 100576610 | mitochondrial ribonuclease P protein 3-like | 172.91161 | -0.4591328 | 0.0643239 | 0.0235932 | -0.0005263 |
| GB40901 | probable cleavage and polyadenylation specificity factor subunit 2 isoform X1 | 172.49223 | -0.0634519 | 0.1359103 | 1.2895750 | -0.0328744 |
| GB44940 | exosome complex exonuclease RRP44-like isoform X1 | 172.44818 | -0.1231613 | 0.0184006 | -0.0184116 | 0.0612947 |
| GB50421 | prosaposin isoformX1 | 172.35206 | -0.1462134 | -0.1051595 | 0.0445204 | -0.1935779 |
| GB50747 | trafficking protein particle complex subunit 13-like | 172.30048 | -0.3015649 | -0.0228584 | 0.0024468 | 0.1165972 |
| GB43820 | PHD finger and CXXC domain-containing protein CG17446-like isoform 1 | 172.08648 | -0.4095687 | -0.0038770 | 0.0133290 | 0.0896747 |
| GB50846 | vacuolar protein-sorting-associated protein 36 | 172.06052 | -0.1934524 | -0.0594418 | 0.0321774 | 0.0339590 |
| GB54227 | phosducin-like protein-like isoform 1 | 171.96010 | -0.1002961 | -0.1224734 | 0.0583757 | 0.1704157 |
| GB48450 | bobby sox, transcript variant X3 | 171.92532 | -0.2119543 | -0.0972506 | 0.0892759 | 0.0377658 |
| GB45905 | thyroid receptor-interacting protein 11 isoform X3 | 171.89349 | -0.4200163 | -0.1624605 | 0.0417834 | -0.0689656 |
| GB51462 | methyltransferase-like protein 23-like isoform X2 | 171.63545 | -0.6825529 | 0.0278093 | 0.4308139 | 0.0774094 |
| GB43305 | ubiquitin specific protease-like | 171.46594 | -0.0817327 | -0.0471341 | -0.0294597 | 0.0644272 |
| GB43888 | parafibromin | 171.40017 | 0.0141943 | 0.1746474 | -0.0095792 | 0.0361076 |
| GB52977 | UBX domain-containing protein 1-A-like | 171.15919 | -0.3004957 | 0.0598596 | -0.0715498 | 0.2815663 |
| GB54938 | ATPase WRNIP1-like isoform X6 | 171.12996 | -0.3365107 | 0.0025946 | 0.0240983 | -0.1116194 |
| GB42479 | T-complex protein 1 subunit eta | 171.02672 | 0.1927516 | 0.1401026 | -0.0157018 | -0.0150165 |
| GB42447 | WW domain-binding protein 11-like | 170.59498 | -0.0671181 | 0.0354914 | 0.0095909 | 0.0582653 |
| GB41300 | bystin isoform 1 | 170.47301 | -0.0852244 | 0.0873668 | -0.0515452 | -0.0350653 |
| GB44865 | rab GTPase-binding effector protein 1-like isoform X2 | 170.16600 | 0.2376208 | 0.1144210 | 0.0558266 | 0.0795817 |
| GB45370 | DNA-directed RNA polymerase I subunit RPA2 isoform X2 | 170.03043 | 0.0207095 | -0.0427381 | -0.4123049 | -0.0850423 |
| GB44414 | alpha-mannosidase 2 isoform X1 | 169.96329 | -0.1246265 | -0.0808181 | 0.0591496 | -0.0464710 |
| GB40463 | ethanolaminephosphotransferase 1-like | 169.76706 | -0.0372503 | 0.0365725 | -0.0518489 | 0.1223341 |
| GB54300 | probable U2 small nuclear ribonucleoprotein A’ isoform X1 | 169.48055 | -0.0599139 | 0.0316384 | -0.0458707 | 0.1001505 |
| GB42451 | serine/threonine-protein kinase TBK1 | 169.37036 | -0.3205615 | 0.1376725 | 0.0047741 | 0.1421056 |
| GB50972 | gastrulation defective protein 1 homolog | 169.31805 | -0.1971693 | 0.0657208 | 0.1903877 | -0.0034529 |
| GB51706 | histone-lysine N-methyltransferase SETD2-like isoform X1 | 169.31045 | -0.4481025 | -0.0162006 | 0.0935864 | 0.1184476 |
| GB54377 | male-specific lethal 1 homolog | 169.30235 | -0.0107089 | 0.0787733 | 0.0791723 | -0.3478216 |
| GB52165 | probable ATP-dependent RNA helicase YTHDC2 isoform X1 | 169.14874 | -0.2166006 | -0.1492968 | -0.0495434 | 0.1044721 |
| GB40320 | protein tumorous imaginal discs, mitochondrial-like isoform X1 | 169.05348 | -0.1619242 | 0.0041297 | 0.0375536 | -0.0424999 |
| GB51655 | nucleoporin GLE1-like | 168.83463 | -0.0847804 | -0.0635058 | -0.0393827 | 0.0140765 |
| GB42997 | uncharacterized protein LOC100578201 | 168.75985 | -0.3800505 | 0.1690311 | 0.1112928 | 0.0019192 |
| GB40563 | threonine–tRNA ligase, cytoplasmic-like isoform X1 | 168.65026 | 0.1893858 | 0.0081525 | -0.0304706 | -0.0437406 |
| GB43815 | probable 39S ribosomal protein L23, mitochondrial-like isoform X1 | 168.32631 | -0.1585833 | -0.0029415 | -0.0209984 | 0.0871642 |
| GB42645 | kinesin B | 168.28858 | -0.5371475 | 0.2744939 | 0.1776832 | -0.0702883 |
| GB42933 | serine/threonine-protein kinase TAO1 isoform X2 | 168.16786 | -0.1176620 | -0.0135005 | -0.1030836 | -0.0665595 |
| GB53934 | rho GTPase-activating protein 24-like | 168.01757 | -0.3242732 | 0.0554416 | -0.0384074 | 0.1041209 |
| GB52990 | BRISC and BRCA1-A complex member 1-like | 167.99128 | -0.3254478 | 0.1174936 | 0.0108915 | 0.0004634 |
| GB42381 | E3 ubiquitin-protein ligase RNF126-like isoform X5 | 167.95791 | -0.0659800 | 0.0049379 | 0.0546401 | 0.0810624 |
| GB55397 | vesicle-associated membrane protein 7 | 167.88748 | 0.1419213 | -0.0867286 | -0.0365991 | 0.0521229 |
| GB53172 | ADAM 17-like protease-like isoform 2 | 167.54930 | 0.0748085 | -0.0450701 | 0.0236531 | 0.1024472 |
| GB44189 | glutamic acid-rich protein-like | 167.43525 | 0.0880143 | 0.0526048 | -0.0479209 | -0.0654774 |
| GB44397 | regulator of microtubule dynamics protein 1-like isoform X1 | 167.40823 | -0.1989022 | -0.0226060 | -0.0122923 | 0.0010582 |
| GB49996 | regulator of G-protein signaling loco isoform X4 | 167.17637 | -0.0093953 | -0.0241350 | 0.0663415 | 0.1139634 |
| GB50589 | DDRGK domain-containing protein 1-like | 166.95040 | -0.1019996 | -0.0952753 | -0.0483397 | 0.0635181 |
| GB53147 | cell division cycle protein 16 homolog isoform X2 | 166.63509 | -0.1296634 | 0.1834347 | 0.1533974 | 0.0234167 |
| GB47744 | sorting nexin-25-like | 166.61334 | -0.0488921 | -0.0729742 | 0.1539323 | 0.1408092 |
| GB47826 | alkylated DNA repair protein alkB homolog 8-like | 166.42901 | -0.5753528 | -0.0646846 | -0.0830894 | 0.0462685 |
| GB44104 | V-type proton ATPase subunit G | 166.40192 | 0.0170920 | -0.0168743 | 0.0833584 | 0.0707069 |
| GB40780 | zinc finger protein 330 homolog | 166.14575 | -0.4061068 | 0.0770956 | 0.0679618 | 0.0914965 |
| GB50183 | cylicin-2-like | 166.13693 | 0.1655849 | 0.0167800 | 0.2181523 | -0.1644786 |
| GB51867 | ubiquitin carboxyl-terminal hydrolase isozyme L5 | 166.11528 | -0.0131991 | 0.0253632 | -0.0453921 | 0.1260061 |
| GB53132 | trifunctional enzyme subunit beta, mitochondrial-like | 166.07786 | 0.2534760 | -0.0294923 | -0.1399508 | 0.1851779 |
| GB41755 | vacuolar protein sorting-associated protein 37A | 165.53325 | 0.1859728 | 0.0709252 | 0.0751567 | -0.0271144 |
| GB48266 | Hermansky-Pudlak syndrome 3 protein homolog | 165.22780 | -0.1460207 | -0.0912595 | 0.1312040 | 0.1240288 |
| GB51701 | CUE domain-containing protein 1 | 165.13770 | -0.2843529 | -0.2020643 | -0.0828381 | 0.1441842 |
| GB49608 | protein angel-like isoform X1 | 164.99695 | -0.4745777 | -0.1501227 | 0.0097089 | -0.3911702 |
| GB53228 | vacuolar protein sorting-associated protein 37B-like | 164.95976 | -0.1001412 | 0.0324804 | 0.0516558 | 0.0672719 |
| GB45119 | elongation factor Ts, mitochondrial-like | 164.90078 | 0.4157426 | -0.0401995 | 0.0311234 | -0.0351009 |
| GB45360 | exportin-2 | 164.63346 | 0.3306912 | 0.1146233 | -0.0533202 | 0.0391325 |
| GB49226 | tetratricopeptide repeat protein 1-like | 164.56124 | -0.1312727 | 0.0647437 | -0.0042155 | 0.1193453 |
| GB50920 | 39S ribosomal protein L37, mitochondrial | 164.53509 | -0.0627923 | 0.0821423 | -0.0219706 | 0.0439987 |
| GB54957 | anaphase-promoting complex subunit 7 isoform X1 | 164.37643 | 0.1083459 | -0.0770647 | 0.0286664 | -0.0151621 |
| GB55017 | putative GTP-binding protein 6-like isoform X1 | 164.24508 | -0.4279412 | 0.0980476 | 0.2020634 | 0.0654012 |
| GB42207 | eukaryotic translation initiation factor 2D-like isoform X2 | 164.23052 | -0.4402548 | -0.1878352 | 0.0848978 | 0.0844453 |
| GB48145 | tyrosine-protein phosphatase non-receptor type 61F-like | 164.16294 | -0.3001985 | 0.3129825 | -0.0224264 | 0.3619577 |
| GB44883 | protein bunched, class 2/F/G isoform isoform X2 | 164.08452 | 0.1423153 | 0.0431774 | 0.1096459 | 0.0820739 |
| GB54809 | cell wall protein IFF6-like isoform X1 | 164.07411 | -0.2793603 | 0.0938404 | 0.0189103 | -0.0487562 |
| GB46339 | heat shock protein 75 kDa, mitochondrial isoform 1 | 163.97346 | 0.2227431 | 0.0858850 | 0.0825314 | 0.1098457 |
| 102654871 | DCN1-like protein 3-like | 163.73872 | -0.5446257 | -0.1149403 | -0.0529742 | -0.0266523 |
| GB44262 | spectrin beta chain, non-erythrocytic 5 isoform X4 | 163.63779 | -0.0348910 | -0.0986757 | -0.1006847 | -0.0922269 |
| GB54321 | RuvB-like 2 isoform X1 | 163.57363 | 0.2691298 | 0.0885141 | 0.1077559 | -0.0199705 |
| GB13213 | eukaryotic translation initiation factor 4 gamma | 163.51563 | -0.3398430 | 0.0132426 | -0.0498854 | 0.0745579 |
| GB52916 | phosphatidylinositide phosphatase SAC2-like isoform X2 | 163.51474 | -0.2041823 | 0.0025506 | 0.1583330 | 0.1866605 |
| GB51426 | asparagine synthetase domain-containing protein 1 isoform X6 | 163.37313 | -0.1068022 | -0.1522976 | 0.0818879 | 0.0731652 |
| GB44028 | vacuolar protein sorting-associated protein 35 isoform X2 | 163.27214 | -0.0579076 | -0.3461417 | 0.0382400 | -0.0489067 |
| GB45341 | polypeptide N-acetylgalactosaminyltransferase 35A-like | 163.09388 | -0.4167607 | 0.0881717 | 1.6656021 | -0.0117249 |
| GB54701 | suppressor of G2 allele of SKP1 homolog isoformX1 | 162.66754 | -0.1767265 | 0.1201854 | -0.0086041 | 1.0849255 |
| GB42667 | trafficking protein particle complex subunit 4-like | 162.08771 | -0.2197603 | -0.1750175 | 0.0386779 | -0.0055323 |
| GB41333 | DNA-directed RNA polymerase III subunit RPC1-like isoform X1 | 161.61636 | -0.1577234 | 0.1680904 | -0.1650504 | -0.3751015 |
| GB54198 | liprin-alpha-2-like isoform X13 | 161.55086 | -0.1542755 | -0.0349291 | 0.3996912 | 0.0008822 |
| GB40399 | synaptobrevin homolog YKT6 isoformX1 | 161.40256 | -0.0445405 | 0.0481756 | 0.0212926 | -0.0047779 |
| GB44449 | putative 28S ribosomal protein S5, mitochondrial isoform X1 | 161.33064 | 0.1662338 | -0.0282685 | 0.0311763 | 0.0062591 |
| GB44293 | lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 | 161.00406 | -0.3920475 | -0.0260276 | 0.0587180 | -0.0768334 |
| GB55595 | probable 39S ribosomal protein L24, mitochondrial | 160.66970 | -0.2371454 | -0.0241757 | 0.0492170 | 0.0447080 |
| GB49250 | heme oxygenase isoform X1 | 160.54774 | -0.7061596 | -0.1862855 | 0.0368602 | 0.0448400 |
| GB47236 | micronuclear linker histone polyprotein isoform X1 | 160.44234 | -0.0470159 | 0.0382495 | 0.0640319 | 0.0621723 |
| GB49158 | dynactin subunit 2, transcript variant X2 | 159.93667 | -0.0774768 | 0.0718003 | -0.2085030 | 0.1296526 |
| GB47884 | TELO2-interacting protein 2-like isoform X1 | 159.81577 | -0.2698585 | 0.3018826 | 0.1126823 | 0.1121113 |
| GB55913 | integrator complex subunit 2 | 159.76275 | -0.3696034 | 0.0859222 | 0.0421902 | -0.0477617 |
| GB46147 | uncharacterized protein LOC408724 | 159.23819 | 0.1867932 | -0.0042754 | -0.0177846 | 0.0331580 |
| GB51413 | C3 and PZP-like alpha-2-macroglobulin domain-containing protein 8-like | 159.11460 | 0.1038200 | 0.0239723 | 0.1542679 | 0.0536660 |
| GB55847 | ubiquitin activating enzyme 1 isoform 1 | 159.07330 | -0.0976432 | -0.0081211 | -0.0101689 | -0.2960334 |
| GB44597 | pleiotropic regulator 1 | 159.03511 | 0.1123349 | 0.0778913 | 0.1442624 | 0.2046111 |
| GB40881 | transcription initiation factor IIA subunit 1 isoform 1 | 159.01949 | -0.3585375 | 0.0154090 | -0.0721560 | -0.0224567 |
| GB41600 | high mobility group protein 20A-like isoform X1 | 158.97672 | -0.7114929 | 0.1535458 | -0.0759391 | 0.0543183 |
| GB45297 | zinc finger FYVE domain-containing protein 1-like | 158.81251 | 0.1142162 | 0.0398985 | 0.0831260 | 0.0221022 |
| GB51400 | probable phenylalanine–tRNA ligase alpha subunit-like isoform X3 | 158.77272 | 0.1149628 | 0.0309227 | 0.0123511 | 0.1653306 |
| GB49652 | cell division cycle 5-like protein-like | 158.66415 | -0.2428431 | 0.0993961 | 0.0950867 | 1.2396760 |
| GB48349 | zinc finger CCCH domain-containing protein 18-like isoform X3 | 158.53066 | -0.5376290 | 0.0561838 | 0.0687067 | 0.0553108 |
| GB44901 | conserved oligomeric Golgi complex subunit 2-like isoform X2 | 158.42607 | -0.2183987 | -0.0307229 | 0.0649127 | 0.1440679 |
| GB47434 | F-box only protein 6-like isoform 1 | 158.21233 | -0.0175862 | -0.0523391 | 0.0833399 | 0.0712098 |
| GB45822 | corepressor interacting with RBPJ 1-like | 158.19150 | -0.2960770 | 0.1024649 | 0.1067356 | 0.1091841 |
| GB48534 | probable protein phosphatase 2C T23F11.1-like isoform X1 | 158.18279 | -0.1736775 | 0.0337026 | 0.0297280 | 0.0012795 |
| GB40760 | syntaxin-5-like | 158.15837 | 0.0685463 | 0.0105051 | -0.0527290 | 0.0229458 |
| GB43848 | glucose-induced degradation protein 8 homolog isoform X2 | 157.96106 | 0.2763054 | 0.1174265 | 0.1410859 | 0.0526052 |
| GB54244 | drebrin-like protein-like | 157.85489 | -0.1819463 | 0.0939261 | 0.1725097 | -0.0361985 |
| GB52462 | THUMP domain-containing protein 3-like isoform X1 | 157.78967 | -0.5044879 | 0.0216561 | -0.2131150 | 0.1666244 |
| GB40672 | type II inositol 1,4,5-trisphosphate 5-phosphatase-like isoform X1 | 157.69281 | -0.1514453 | -0.0449708 | 0.0888520 | 0.1930789 |
| GB49112 | probable uridine-cytidine kinase-like isoformX1 | 157.37727 | 0.1936417 | 0.0438149 | 0.1423841 | -0.0199275 |
| GB52060 | septin-1-like | 157.24552 | -0.3205289 | 0.0598405 | 0.0777303 | 0.0692801 |
| GB49020 | DNA-directed RNA polymerase III subunit RPC5 | 157.22864 | -0.3737857 | 0.3001013 | -0.6084797 | 0.1539672 |
| GB42771 | F-box/WD repeat-containing protein 5-like | 157.09497 | -0.6953162 | 0.3075574 | -0.0551481 | 0.1835573 |
| GB43266 | C-terminal-binding protein isoform X3 | 157.01081 | -0.3956263 | -0.1829912 | -0.0647185 | -0.0610275 |
| GB40833 | UPF0396 protein CG6066-like isoform X3 | 156.97193 | -0.2613537 | -0.0159997 | -0.0295758 | 0.0235516 |
| GB54570 | golgin subfamily A member 2-like isoform X2 | 156.96687 | -0.3626323 | 0.0492098 | 0.0256836 | 0.1067349 |
| 725816 | homocysteine S-methyltransferase 2-like | 156.84552 | -0.2766233 | -0.2226405 | -0.0490383 | -0.0094930 |
| GB49537 | probable DNA mismatch repair protein Msh6 | 156.72155 | -0.0913736 | -0.0060320 | -0.0873948 | -0.0262102 |
| GB42564 | serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform-like isoform X2 | 156.49222 | -0.0977575 | 0.0563203 | 0.1585262 | 0.1125627 |
| GB45761 | T-complex protein 1 subunit gamma | 156.21079 | 0.4197650 | 0.0829152 | 0.3256464 | 0.4526983 |
| GB46115 | U4/U6 small nuclear ribonucleoprotein Prp31-like isoform X2 | 156.19634 | 0.2545786 | 0.0692318 | 0.0589038 | -0.0514496 |
| GB55994 | dual specificity protein phosphatase 12 isoform X2 | 155.99166 | -0.1669523 | -0.0263361 | 0.0072606 | -0.0072627 |
| GB46248 | sorting nexin lst-4-like isoform 1 | 155.91985 | 0.0553885 | -0.0298433 | 0.1663085 | -0.1187090 |
| GB53317 | dentin sialophosphoprotein-like | 155.90410 | -0.6522746 | 0.0964948 | 0.0428159 | 0.0660586 |
| GB51762 | THO complex subunit 5 homolog isoform X1 | 155.63752 | 0.0519010 | 0.1764810 | 0.0305450 | 0.0049660 |
| GB48817 | nuclear protein localization protein 4 homolog isoform X3 | 155.63351 | 0.3510511 | -0.0333526 | 0.1294790 | 0.2191341 |
| GB43502 | E3 UFM1-protein ligase 1 homolog | 155.52510 | -0.3546514 | 0.1505166 | -0.0282551 | 0.0438175 |
| GB44289 | ataxin-3-like isoform X4 | 155.44275 | -0.1315793 | -0.0400609 | 0.8082165 | 0.0414597 |
| GB40308 | hsp70-binding protein 1-like | 155.24348 | 0.3304605 | 0.0523219 | 0.6874181 | 0.0240131 |
| GB45397 | MATH and LRR domain-containing protein PFE0570w-like | 155.16681 | -0.7159460 | 0.1353533 | 0.0122866 | 0.1029342 |
| GB46459 | islet cell autoantigen 1-like isoform X2 | 155.08611 | -0.0727968 | 0.1408801 | -0.0120188 | 0.1753914 |
| GB41802 | uncharacterized protein LOC552003 isoform X4 | 154.88251 | 0.0770305 | 0.0644961 | -0.0174025 | -0.0037769 |
| GB48821 | structural maintenance of chromosomes protein 6-like isoform X4 | 154.72081 | -0.2476209 | 0.0848117 | 0.0222281 | 0.0855998 |
| GB47184 | SCY1-like protein 2-like | 154.64724 | -0.2355251 | -0.0855877 | 0.7382324 | 0.0124713 |
| GB50997 | structural maintenance of chromosomes protein 3 | 154.64304 | 0.0420086 | -0.0540943 | -0.0917712 | 0.0668986 |
| GB42274 | regulation of nuclear pre-mRNA domain-containing protein 1B-like isoform X2 | 154.54454 | 0.2251752 | 0.0585259 | 0.0854331 | 0.0121938 |
| GB51564 | prefoldin subunit 3-like | 154.53343 | 0.2027363 | -0.0077751 | -0.0360888 | 0.0573586 |
| GB43955 | protein PFF0380w-like | 154.49141 | 0.0415650 | -0.1188688 | 0.1430416 | 0.0099351 |
| GB40388 | exocyst complex component 8 | 154.44665 | 0.0933704 | 0.0924731 | 0.0685702 | 0.0059113 |
| GB51586 | 26S protease regulatory subunit 10B | 154.42934 | 0.0638296 | -0.0472381 | -0.1868630 | 0.0377330 |
| GB41759 | N-acetylglucosaminyl-phosphatidylinositol biosynthetic protein-like isoform X2 | 154.29385 | -0.4298497 | -0.1114510 | -0.0285141 | -0.0119614 |
| GB51603 | peptidyl-alpha-hydroxyglycine alpha-amidating lyase 1-like isoform X5 | 154.19915 | -0.6376376 | 0.0434204 | -0.0214823 | 0.1321119 |
| GB50511 | DNA mismatch repair protein Mlh1 isoform X2 | 154.04259 | -0.0419613 | 0.0014262 | -0.0192785 | 0.1477541 |
| GB50998 | pre-mRNA 3’-end-processing factor FIP1-like | 153.79484 | 0.3518992 | 0.0668569 | 0.0088979 | 0.0197453 |
| GB40364 | conserved oligomeric Golgi complex subunit 8-like | 153.74298 | -0.3885971 | 0.0381729 | 0.0021637 | 0.0696396 |
| GB41852 | centromere-associated protein E isoform X8 | 153.34829 | -0.0967937 | 0.0656524 | 0.6213701 | 0.0776662 |
| GB45544 | uncharacterized protein LOC552484 | 153.29865 | -0.1977929 | -0.0776993 | -0.1765297 | 0.1469585 |
| GB55753 | dehydrogenase/reductase SDR family protein 7-like isoform 1 | 153.26811 | -0.1757835 | -0.0158448 | -0.0351785 | 0.1077771 |
| GB45693 | DNA topoisomerase 3-beta-1-like isoform 2 | 153.20023 | 0.1006986 | 0.1061570 | 0.1217369 | 0.0258593 |
| GB51599 | collagen type IV alpha-3-binding protein-like isoformX1 | 153.00299 | 0.0280506 | 0.1170853 | -0.0709939 | 0.0024371 |
| GB48596 | anaphase-promoting complex subunit 4-like isoformX2 | 152.94626 | -0.1827155 | 0.0074676 | 0.1614664 | 0.0395005 |
| GB53191 | arf-GAP domain and FG repeat-containing protein 1 isoform X1 | 152.92572 | -0.1168342 | -0.1267303 | -0.0484076 | -0.0280372 |
| GB44757 | actin-interacting protein 1 isoform X2 | 152.86778 | 0.0624740 | -0.0836119 | 0.0930331 | -0.0178032 |
| GB49773 | sequestosome-1 | 152.77910 | -0.0958243 | -0.2694184 | 0.1680133 | 0.3703431 |
| GB51334 | methionine aminopeptidase 2 | 152.76213 | 0.1680092 | 0.0257564 | 0.0797106 | 0.0272008 |
| GB51968 | conserved oligomeric Golgi complex subunit 3 | 152.69056 | -0.2523187 | 0.0321808 | 0.1223475 | 0.1766898 |
| GB50357 | clathrin heavy chain-like isoform 1 | 152.65882 | -0.3565829 | -0.0349857 | -0.0290474 | -0.4245458 |
| GB41668 | serine/threonine-protein kinase/endoribonuclease IRE1 isoform X2 | 152.62752 | -0.2610915 | -0.0394913 | 0.1167377 | 0.1303052 |
| GB48955 | microfibrillar-associated protein 1 | 152.62088 | -0.0027927 | 0.0980686 | -0.0238326 | 0.0008515 |
| GB47180 | TATA-box-binding protein-like isoform 1 | 152.57215 | -0.5556062 | 0.0137772 | -0.1563645 | 0.1407033 |
| GB50280 | iron-sulfur cluster assembly 2 homolog, mitochondrial-like | 152.51442 | -0.1773518 | -0.2051342 | -0.0224675 | 0.0686623 |
| 102656136 | prefoldin subunit 4-like | 152.48244 | -0.6925061 | 0.1097711 | -0.0739651 | 0.1990536 |
| GB42874 | transforming acidic coiled-coil-containing protein 3 isoform X1 | 152.26682 | -0.2456148 | 0.1220464 | 0.1710556 | 0.1332140 |
| GB50104 | KAT8 regulatory NSL complex subunit 3 isoform X2 | 152.23090 | -0.3333269 | 0.1260357 | 0.5875565 | 0.1923016 |
| GB48855 | uncharacterized protein YJR142W-like | 152.19894 | -0.1858051 | 0.0098299 | -0.0879237 | -0.0623119 |
| GB44338 | small G protein signaling modulator 3 homolog | 151.68939 | 0.1253131 | 0.0098320 | 0.0499533 | 0.1876984 |
| GB54685 | uncharacterized protein C3orf18 homolog isoform X1 | 151.66606 | -0.2094785 | -0.1600359 | 0.1588197 | 0.0112474 |
| GB46457 | GPI ethanolamine phosphate transferase 3-like isoform X2 | 151.46086 | -0.0785608 | -0.0716776 | 0.1152346 | 0.0562230 |
| 411727 | bromodomain-containing protein 8 | 151.38352 | 0.0527861 | 0.1551509 | -0.0768800 | -0.0237612 |
| GB54643 | putative mitochondrial inner membrane protein-like isoform X7 | 151.20165 | -0.2278187 | -0.0243459 | 0.0621358 | -0.0305928 |
| GB49025 | zinc transporter foi-like isoform X4 | 151.02191 | -0.1070890 | -0.1265826 | -0.0467670 | 0.0063989 |
| GB51103 | COP9 signalosome complex subunit 6-like isoform X2 | 150.82226 | 0.1665124 | 0.0855128 | -0.0004730 | 0.0698093 |
| GB42880 | protein FAM8A1-like isoform X2 | 150.73019 | 0.1249839 | -0.1755929 | 0.1332146 | 0.0905954 |
| GB46641 | E3 ubiquitin-protein ligase LRSAM1-like isoform X3 | 150.53735 | -0.3102017 | -0.0741280 | -0.0623466 | 0.0020677 |
| GB41835 | sorting nexin-13-like isoform X2 | 150.32661 | -0.2094181 | -0.0066059 | 0.1205814 | 0.0803841 |
| GB45156 | WD repeat-containing protein 59-like isoform X2 | 150.24606 | 0.0842779 | -0.0722461 | 0.1937462 | -0.0092339 |
| GB43439 | UV radiation resistance-associated gene protein-like | 150.15100 | -0.0118453 | -0.3076536 | 1.4722603 | -0.0045068 |
| GB43470 | regulator of nonsense transcripts 2 | 150.08531 | -0.3128759 | -0.0466710 | -0.0553300 | 0.0013376 |
| GB43215 | actin-related protein 6-like isoform X2 | 149.65414 | 0.3061957 | 0.0777613 | 0.0429150 | 0.0624302 |
| GB51005 | N-alpha-acetyltransferase 20-like isoform X2 | 149.61834 | 0.0149843 | 0.0425788 | 0.1707903 | 0.0985047 |
| GB50360 | zinc finger CCCH domain-containing protein 11A-like | 149.26448 | -0.5424708 | -0.0651305 | 0.1221017 | -0.1112584 |
| GB41340 | uncharacterized protein LOC100579034 isoform X2 | 149.23342 | -0.2945055 | 0.1383336 | -0.0341506 | -0.0166941 |
| GB55701 | putative aldehyde dehydrogenase family 7 member A1 homolog isoform 2 | 149.10584 | 0.5246497 | -0.0914360 | 0.0393989 | -0.1075361 |
| GB51539 | 28S ribosomal protein S17, mitochondrial isoform X2 | 149.10008 | -0.1191544 | -0.0634678 | -0.0343159 | 0.0679068 |
| GB50658 | DNA primase large subunit | 148.85628 | -0.0074660 | 0.1970793 | -0.0851038 | 0.2426718 |
| GB53251 | N-alpha-acetyltransferase 25, NatB auxiliary subunit isoform X2 | 148.83651 | 0.0016318 | 0.1537186 | -0.9775687 | 0.0615761 |
| GB43300 | RNA-binding protein squid-like isoform X6 | 148.82089 | -0.1284436 | 0.0896448 | 0.0693043 | 0.0977145 |
| GB50482 | uncharacterized protein LOC724971 isoform X2 | 148.54867 | -0.0441563 | -0.2396737 | 0.0017989 | 0.0863729 |
| GB54657 | Rad54 protein isoform X2 | 148.45257 | -0.2084683 | -0.1783769 | 0.1011935 | 0.1676428 |
| GB52541 | probable cation-transporting ATPase 13A1-like | 148.28464 | -0.2102158 | 0.1385270 | -0.0268470 | -0.0112677 |
| GB46262 | IQ motif and SEC7 domain-containing protein 2 isoform X5 | 148.15532 | -0.5758259 | -0.0000689 | 0.0686664 | 0.0098451 |
| GB53005 | gem-associated protein 8-like | 147.97037 | 0.0452613 | -0.0116292 | -0.0571413 | 0.0730754 |
| GB50935 | mitochondrial chaperone BCS1-like | 147.84204 | -0.1330652 | -0.0449103 | -0.0329387 | 0.1282745 |
| GB51271 | protein Peter pan-like | 147.71876 | -0.2767305 | 0.0936454 | 0.1938348 | 0.1412980 |
| GB52980 | uncharacterized protein LOC552428 | 147.64472 | -0.0292196 | -0.0688415 | 0.0505954 | -0.0105940 |
| GB45566 | protein EFR3 homolog cmp44E isoformX1 | 147.60468 | 0.0331860 | 0.0490985 | 0.1458039 | 0.2391108 |
| GB44347 | TBC1 domain family member 16 isoform X2 | 147.58413 | -0.1259259 | -0.1033395 | 0.2509934 | 0.1114623 |
| GB49949 | WD repeat domain phosphoinositide-interacting protein 2-like isoform X2 | 147.45558 | -0.0129942 | 0.0940478 | 0.1343766 | -0.0402042 |
| GB51634 | glucose 1,6-bisphosphate synthase isoform X2 | 147.31216 | 0.1965173 | -0.0635826 | -0.8589679 | 0.1254671 |
| GB50078 | ATP-dependent RNA helicase abstrakt | 147.27515 | -0.0003280 | 0.1434898 | 0.0536618 | 0.0638111 |
| GB54909 | dynamin-binding protein-like isoform X2 | 147.18917 | -0.0634231 | -0.0129333 | 0.1047481 | 0.0360023 |
| GB40113 | UBX domain-containing protein 4-like isoform 1 | 147.05676 | -0.2843523 | -0.0303492 | -0.0995521 | 0.0366073 |
| GB43858 | INO80 complex subunit E-like | 147.04654 | -0.1072097 | 0.0330119 | 0.0180872 | -0.0034026 |
| GB47748 | two pore calcium channel protein 1 | 146.92411 | -0.3347448 | -0.1533686 | -0.0003191 | 0.0804698 |
| GB53915 | dimethyladenosine transferase 1, mitochondrial | 146.91968 | -0.5044836 | -0.0736598 | 0.0740909 | 0.0320441 |
| GB43625 | RNA methyltransferase-like protein 1-like | 146.91910 | -0.2491927 | 0.1389611 | -0.1140519 | 0.0333435 |
| GB49161 | probable aminoacyl tRNA synthase complex-interacting multifunctional protein 2 isoform X2 | 146.89512 | -0.5497647 | -0.0340830 | 0.0525094 | 0.0599583 |
| GB47812 | LOW QUALITY PROTEIN: regulatory-associated protein of mTOR | 146.88865 | -0.1627796 | 0.0183474 | 0.0638252 | -0.1182190 |
| GB46894 | pre-mRNA-splicing factor SYF1-like isoformX1 | 146.86414 | -0.0288915 | 0.2438213 | -0.0450779 | 0.1762920 |
| GB43553 | 5’-AMP-activated protein kinase catalytic subunit alpha-2 isoform X3 | 146.69480 | -0.1601746 | -0.0069763 | 0.1227680 | 0.0816390 |
| GB49789 | 28S ribosomal protein S29, mitochondrial isoformX1 | 146.63563 | -0.6207520 | -0.0201670 | 0.3400965 | 0.1896255 |
| GB49570 | centaurin-gamma-1A | 146.57702 | -0.1922250 | -0.0506733 | 0.1274670 | 0.1426742 |
| GB53650 | glycerol kinase-like isoform X2 | 146.47017 | -0.5182606 | -0.2030319 | -0.0501066 | -0.0264044 |
| GB44176 | integrator complex subunit 8 | 146.40593 | 0.2076989 | -0.0467532 | 0.1181645 | 0.0967430 |
| GB44991 | RNA-binding protein cabeza-like isoform X1 | 146.40230 | -0.0627078 | -0.0382216 | 0.0977350 | -0.0035867 |
| GB45122 | mitochondrial assembly of ribosomal large subunit protein 1-like | 146.39645 | -0.4154315 | 0.0189759 | 0.4910612 | -0.0037883 |
| GB52910 | octopamine receptor | 146.27188 | -0.7733899 | -0.0239961 | -0.8802540 | 0.1595786 |
| GB50740 | ubiquitin-conjugating enzyme E2 S-like isoform X2 | 146.18715 | 0.2638466 | 0.1183353 | -0.5311281 | 0.1588701 |
| GB42216 | hepatoma-derived growth factor-related protein 2-like isoform 2 | 146.16623 | 0.0061614 | 0.0591646 | -0.0399919 | 0.0481260 |
| GB47964 | zinc finger protein 543-like isoform X1 | 146.08906 | 0.2008906 | 0.0143720 | 0.1314465 | 0.0134619 |
| GB43074 | phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform-like isoform X1 | 146.08717 | -0.0840933 | -0.0157862 | 0.1322251 | 0.1335276 |
| GB53359 | alkylated DNA repair protein alkB homolog 1 | 145.95253 | -0.0215505 | 0.0308703 | -0.0410398 | -0.0037074 |
| GB54342 | DEAD-box helicase Dbp80 isoform X2 | 145.75995 | 0.2047489 | 0.0379695 | -0.1070177 | 0.1301033 |
| GB47417 | mediator of RNA polymerase II transcription subunit 15 isoform X2 | 145.74255 | -0.1403195 | -0.0199005 | 0.1320668 | 0.0162830 |
| GB49634 | trafficking protein particle complex subunit 12-like | 145.63119 | -0.0081125 | 0.0771374 | 0.2138523 | 0.0232734 |
| GB43163 | nuclear factor related to kappa-B-binding protein | 145.39973 | -0.2844265 | 0.0198382 | -1.0199566 | -0.0487731 |
| 412933 | geranylgeranyl transferase type-1 subunit beta isoform X1 | 145.14985 | 0.3690186 | -0.0753994 | -0.0119145 | -0.0719928 |
| GB48982 | uncharacterized protein LOC413055 | 145.13952 | -0.2927182 | 0.0080430 | 3.6495997 | -0.0194245 |
| GB40301 | aldose 1-epimerase-like isoform X2 | 145.03562 | -0.1153919 | -0.0107421 | 0.0708060 | 0.0430586 |
| GB18555 | tether containing UBX domain for GLUT4 | 144.69514 | -0.3842693 | 0.0506226 | -0.0527979 | 0.0268992 |
| GB54831 | nuclear transcription factor Y subunit gamma-like isoform X1 | 144.33637 | -0.1201217 | -0.0925929 | 0.0248679 | -0.0984831 |
| GB45689 | mediator of RNA polymerase II transcription subunit 25 isoform X3 | 144.07438 | 0.0177484 | 0.1088169 | -0.0341463 | 0.1157417 |
| GB48625 | probable ATP-dependent RNA helicase spindle-E isoform X2 | 144.01831 | 0.2218261 | 0.1606669 | -0.1110084 | -0.1425290 |
| GB52677 | forkhead box protein K1-like isoform X2 | 143.98187 | 0.0352617 | 0.1499677 | 0.0509449 | 0.0618666 |
| GB50699 | protein SET-like isoform X1 | 143.93542 | 0.4869852 | 0.0395231 | 0.1473844 | 0.1870108 |
| GB45039 | probable RNA polymerase II nuclear localization protein SLC7A6OS-like | 143.90839 | -0.0149136 | 0.2579033 | 0.0955197 | 0.0981995 |
| GB54951 | ubiquitin carboxyl-terminal hydrolase 46-like isoform X1 | 143.78492 | -0.1944345 | 0.1286488 | -0.0317593 | 0.0731775 |
| GB42918 | protein NipSnap | 143.60882 | 0.3893414 | -0.0577951 | 0.0478783 | 0.0116827 |
| GB50069 | vacuolar protein sorting-associated protein 51 homolog | 143.55848 | -0.0767230 | 0.0328014 | 0.0255298 | 0.1752537 |
| GB40556 | WD repeat-containing protein 61-like | 143.50436 | -0.1973982 | -0.0560545 | 0.0836334 | 0.1941122 |
| GB40798 | serine/arginine-rich splicing factor 7 isoform X1 | 143.48383 | 0.1047218 | 0.0635743 | 0.6802646 | 0.0680094 |
| GB52737 | vacuolar protein sorting-associated protein 41 homolog isoform X2 | 143.39837 | 0.0939460 | -0.0513516 | 0.1155147 | -0.0803081 |
| GB45491 | G patch domain and ankyrin repeat-containing protein 1 homolog | 143.39216 | -0.2389827 | -0.0608652 | 0.0935341 | 0.0900960 |
| GB42352 | elongation factor Tu GTP-binding domain-containing protein 1-like, transcript variant X4 | 143.32836 | 0.0370350 | 0.0593796 | 0.1600560 | -0.0692563 |
| GB50688 | 26S proteasome non-ATPase regulatory subunit 7-like | 143.31169 | 0.1646387 | 0.0457201 | -0.0798990 | 0.0488735 |
| GB52904 | nuclear hormone receptor HR96 isoform X2 | 143.15966 | -0.2536658 | -0.0242007 | 0.0607065 | 0.0275286 |
| GB40891 | RING finger protein 157-like isoform X3 | 143.09442 | -0.0127784 | 0.0901122 | -0.0494670 | -0.0032095 |
| GB44930 | ell-associated factor Eaf-like isoform X1 | 143.04323 | 0.0553170 | 0.1431589 | -0.1077929 | 0.0053112 |
| 102656131 | biogenesis of lysosome-related organelles complex 1 subunit 2-like | 142.98791 | -0.0178170 | -0.2128337 | 0.1523093 | -0.1644945 |
| GB44543 | DNA polymerase alpha subunit B isoform X2 | 142.94143 | 0.1369515 | -0.0921261 | 0.0461638 | 0.1117859 |
| GB40452 | brefeldin A-inhibited guanine nucleotide-exchange protein 1 | 142.89718 | -0.2600671 | 0.0619868 | 0.1226856 | -0.0425863 |
| GB43856 | dnaJ homolog subfamily C member 11-like | 142.83204 | 0.1359915 | 0.1060312 | -0.0860241 | 0.0739310 |
| GB42681 | mucin-5AC-like isoform X3 | 142.48311 | -0.2593462 | 0.0943990 | 0.9291975 | 0.0974932 |
| GB51143 | protein downstream neighbor of son homolog | 142.45888 | -0.2280975 | 0.0445856 | -0.0901412 | -0.0230543 |
| GB45656 | AKT-interacting protein-like | 142.29808 | 0.1243698 | -0.1005726 | 0.0969314 | 0.1110155 |
| GB55943 | protein FAM114A2-like | 142.21758 | -0.5633091 | -0.0599928 | -0.2084455 | 0.0300454 |
| GB41485 | WD repeat-containing protein 55 homolog | 142.12523 | -0.4751221 | -0.0070468 | -0.0730057 | 0.1881630 |
| GB55659 | tudor domain-containing protein 3-like isoform X1 | 142.02828 | -0.2515698 | 0.1749982 | 0.0648678 | 0.1302752 |
| GB55811 | protein MTO1 homolog, mitochondrial-like isoform 1 | 141.86744 | -0.1862467 | -0.0084389 | 0.0832206 | 0.0913612 |
| GB54571 | FACT complex subunit Ssrp1 | 141.75476 | -0.3027032 | 0.0893227 | -0.0689431 | 0.0265783 |
| GB48157 | G patch domain-containing protein 1 homolog | 141.73690 | -0.1698054 | 0.2329948 | 0.1652515 | -0.0839484 |
| GB46247 | eukaryotic translation initiation factor 3 subunit H | 141.61545 | 0.0741075 | 0.1212617 | -0.0518053 | -0.0449024 |
| GB53825 | transcription initiation factor TFIID subunit 2 | 141.57402 | -0.7182932 | -0.0102693 | 0.1116532 | 0.0837886 |
| GB51716 | 60S ribosomal protein L23a | 141.37463 | 0.1647361 | 0.0547823 | 0.1178507 | -0.0855422 |
| 726768 | probable ATP-dependent RNA helicase DDX17-like | 141.34400 | -0.3382651 | -0.0398062 | 0.1450902 | -0.0075527 |
| GB53852 | paired amphipathic helix protein Sin3a isoform X3 | 141.33684 | -0.3072927 | -0.1049860 | 0.1996987 | 0.0109255 |
| GB52306 | dnaJ homolog subfamily C member 13 | 141.07433 | 0.0611104 | 0.0051145 | -0.0075039 | 0.1285935 |
| GB46520 | vacuolar protein sorting-associated protein 26B | 140.98508 | -0.1083042 | -0.0980337 | -0.1315788 | -0.0581219 |
| GB48169 | hamartin isoform X3 | 140.91141 | -0.3428835 | 0.0026822 | 0.1086022 | -0.0192266 |
| GB41781 | pontin protein isoform 1 | 140.59011 | 0.3243930 | 0.1723577 | 0.0273185 | 0.0057170 |
| GB42016 | intron-binding protein aquarius | 140.51079 | -0.0689879 | 0.0735160 | -0.0962411 | -0.1483527 |
| GB43219 | uncharacterized protein LOC724290 | 140.45735 | -0.8579989 | 0.1841622 | 0.0959868 | 0.1294831 |
| GB50141 | erythroid differentiation-related factor 1-like isoform X1 | 140.41115 | 0.0528165 | 0.0662757 | 0.0866553 | 0.0042494 |
| GB46363 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)]-like isoform X2 | 140.40774 | -0.2098523 | 0.1851748 | 0.1243856 | -0.0715522 |
| GB46707 | vacuolar protein-sorting-associated protein 25 | 140.38486 | 0.0039507 | -0.0390442 | -0.0600830 | 0.1608908 |
| GB41257 | probable protein phosphatase CG10417-like | 140.19080 | 0.1989829 | 0.0368436 | -0.0800763 | 0.0611307 |
| GB46963 | probable ATP-dependent RNA helicase DHX36-like isoform X2 | 140.06335 | -0.0712383 | 0.0687471 | 0.1500131 | -0.0065126 |
| GB43894 | histone deacetylase 3 isoform 1 | 139.86318 | -0.2618239 | 0.1149721 | 0.0731897 | -0.1209108 |
| GB49666 | dnaJ homolog subfamily C member 8-like | 139.61783 | -0.5588177 | 0.0518844 | 0.1170377 | 0.0995899 |
| GB53001 | uncharacterized protein LOC100577272 | 139.58343 | -0.0597428 | -0.0228887 | 0.0870111 | 0.1716861 |
| GB47260 | zinc finger protein 598-like isoform X2 | 139.53493 | -0.6018072 | 0.0593800 | -0.1254441 | 0.0124439 |
| GB42230 | protein IWS1 homolog isoformX2 | 139.49157 | -0.0984320 | 0.1884444 | -0.0667762 | 0.0616872 |
| GB51525 | synergin gamma-like | 139.42183 | -0.2746408 | -0.0722776 | -0.0512115 | 0.0874815 |
| GB48115 | major facilitator superfamily domain-containing protein 6 | 139.37720 | 0.1816722 | 0.0090223 | 0.0738597 | 0.0302290 |
| GB52726 | ras association domain-containing protein 8-like isoform X2 | 139.26051 | 0.0672726 | -0.0784122 | 0.0317955 | -0.0357702 |
| GB49089 | integrator complex subunit 4-like | 139.21283 | 0.0066752 | 0.2003307 | -0.0821244 | -0.0356783 |
| GB53333 | V-type proton ATPase catalytic subunit A-like isoform X3 | 139.13542 | 0.0631321 | -0.0582681 | 0.0353159 | 0.0421506 |
| GB51071 | putative deoxyribose-phosphate aldolase-like isoform X2 | 139.08311 | -0.1521037 | -0.0561238 | -0.0358800 | -0.0990012 |
| GB47882 | abl interactor 2 | 139.08188 | 0.1825083 | 0.0552081 | 0.1905020 | -0.0448790 |
| GB53177 | GPN-loop GTPase 1-like | 138.83423 | -0.3927197 | -0.0374977 | 0.0084195 | 0.0631964 |
| GB43186 | symplekin | 138.78656 | 0.0341178 | 0.0946601 | 0.0196254 | -0.0422572 |
| GB48845 | probable rRNA-processing protein EBP2 homolog | 138.65984 | -0.4818392 | 0.1313151 | -0.0786484 | 0.1150881 |
| GB53239 | HEAT repeat-containing protein 6-like | 138.65776 | -0.1728326 | -0.0414187 | -0.1304882 | 0.3312672 |
| GB45173 | autism susceptibility gene 2 protein-like isoform X2 | 138.64028 | -0.2358246 | -0.0024656 | 0.0992694 | 0.0237323 |
| GB52774 | glycylpeptide N-tetradecanoyltransferase 1 | 138.58069 | 0.3960601 | -0.0132580 | 0.0695643 | 0.0267570 |
| GB49612 | protein real-time-like isoform X1 | 138.47267 | -0.0728927 | -0.0563157 | 0.0365994 | -0.0488433 |
| GB44524 | peptidyl-prolyl cis-trans isomerase-like 2-like | 138.42912 | -0.1707275 | 0.1292449 | 0.0165412 | 0.0015772 |
| GB47813 | malonyl-CoA decarboxylase, mitochondrial-like | 138.41601 | -0.5404035 | -0.1220999 | 0.0331273 | 0.0514485 |
| GB51822 | Sip1/TFIP11 interacting protein | 138.41246 | 0.0817711 | 0.1136980 | 0.0002213 | 0.1499593 |
| GB50347 | LOW QUALITY PROTEIN: polymerase delta-interacting protein 2-like | 138.21930 | -0.0506864 | 0.0626628 | 0.1180547 | 0.1184361 |
| GB54684 | ADIPOR-like receptor CG5315-like isoform X2 | 138.16573 | -0.0132813 | -0.1170597 | -0.1006323 | 0.8076149 |
| GB44103 | protein LTV1 homolog | 137.90352 | -0.7821288 | 0.1198173 | -0.0563213 | 0.1690460 |
| GB41865 | DNA-binding protein Ets97D homolog | 137.82075 | 0.1389374 | 0.1294199 | -0.0633536 | 0.1389068 |
| GB43486 | uncharacterized protein LOC552788 | 137.74232 | -0.2052910 | -0.0581209 | 0.1794974 | 0.1533206 |
| GB53328 | DNA/RNA-binding protein KIN17 | 137.72539 | -0.2452054 | -0.2897766 | 0.0707161 | 0.0805750 |
| GB42234 | cat eye syndrome critical region protein 5-like isoform X2 | 137.71571 | -0.4310357 | -0.0670673 | -0.0881225 | 0.0708768 |
| GB40801 | thioredoxin-like protein 4A-like isoform X1 | 137.66692 | 0.0477766 | 0.0464143 | 0.1761375 | 0.1417084 |
| GB40473 | cytoplasmic phosphatidylinositol transfer protein 1 | 137.65860 | 0.0710352 | -0.0610871 | 0.3052914 | 0.0482978 |
| 102655259 | 5-methylcytosine rRNA methyltransferase NSUN4-like isoform X1 | 137.64837 | -0.6670513 | -0.0434200 | -0.6368044 | 0.1138913 |
| GB48928 | MATH and LRR domain-containing protein PFE0570w-like | 137.61629 | 0.0546139 | 0.1549539 | -0.5727548 | 0.0793314 |
| GB40745 | synaptosomal-associated protein 29 | 137.44055 | -0.3484482 | 0.0725336 | 0.0830910 | 0.0384574 |
| GB43288 | ribonuclease P protein subunit p21-like | 137.40079 | 0.1906509 | -0.0044491 | 0.8098851 | 0.1667594 |
| GB54916 | protein Mo25-like isoform X3 | 137.36188 | 0.2550756 | -0.0265836 | 0.1652391 | -0.3404477 |
| GB46909 | basic proline-rich protein-like isoform X2 | 137.23716 | -0.2123446 | 0.1059261 | 0.0718601 | -0.0646851 |
| GB49643 | probable enoyl-CoA hydratase, mitochondrial-like | 137.23292 | 0.2476690 | 0.0166092 | -0.0869911 | -0.0797591 |
| GB54220 | nuclear distribution protein nudE-like 1-A-like isoform X1 | 137.19736 | -0.2308707 | -0.0032020 | 0.1642344 | 0.0979594 |
| GB46016 | transmembrane protein 131-like | 136.87218 | 0.1101604 | -0.0054753 | 0.1260409 | 0.0388035 |
| GB46911 | programmed cell death protein 10-like | 136.85740 | 0.0806119 | 0.0052822 | 0.0121135 | 0.1169640 |
| GB52852 | zinc finger protein 879-like isoform X2 | 136.81537 | -0.1680960 | 0.0089628 | 0.0335278 | 0.0595537 |
| GB52080 | probable GDP-L-fucose synthase isoform X2 | 136.78816 | 0.0081506 | 0.0794921 | 0.1004142 | 0.0757517 |
| GB44642 | F-box/WD repeat-containing protein 9 isoform 1 | 136.75959 | 0.0553680 | -0.1161027 | -0.0225266 | 0.0540146 |
| GB41769 | actin-related protein 8 isoform 1 | 136.72055 | 0.3118071 | 0.0723317 | -0.0062727 | 0.0917738 |
| GB45357 | protein EMSY-like isoform X2 | 136.70638 | -0.5059609 | 0.1394664 | 0.0801023 | 0.1536270 |
| 725378 | nucleolar GTP-binding protein 2 | 136.55027 | -0.3873532 | 0.1390424 | -0.1430756 | 0.0326776 |
| GB41444 | N-acetyltransferase 10-like | 136.50568 | -0.6264784 | 0.0332903 | 0.0665180 | 0.0376139 |
| GB51692 | mediator of RNA polymerase II transcription subunit 14 isoform X2 | 136.45836 | 0.0127212 | -0.0776514 | 0.0781660 | 0.5180514 |
| GB48402 | zinc finger C4H2 domain-containing protein-like isoform X3 | 136.34513 | 0.2716056 | -0.0133514 | 0.0220623 | 0.0991926 |
| GB49491 | anaphase-promoting complex subunit 1 | 136.27749 | -0.4211302 | -0.0144076 | 0.1548942 | 0.2409880 |
| GB53777 | beta-1,4-galactosyltransferase 7 isoform X2 | 136.13953 | 0.0064604 | -0.1413431 | 0.1130475 | 0.1508595 |
| GB53859 | arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2 isoform X1 | 136.12077 | -0.1800735 | 0.0588415 | 0.0805883 | -0.0154329 |
| GB41711 | uncharacterized protein LOC408493 | 136.08265 | -0.2506783 | 0.0776116 | 0.1180021 | 0.0489960 |
| GB48781 | nuclear nucleic acid-binding protein C1D-like isoformX2 | 136.02229 | -0.2442126 | 0.0952193 | 0.0736146 | 0.0387665 |
| GB44345 | non-histone protein 10-like | 135.77585 | -0.2042073 | 0.3226273 | -0.0129531 | -0.0190337 |
| GB51123 | UPF0047 protein C4A8.02c-like | 135.66717 | 0.0511428 | 0.0026482 | 0.0679195 | -0.0067162 |
| GB53236 | retrograde Golgi transport protein RGP1 homolog isoform X1 | 135.63921 | -0.2047676 | 0.0065631 | -0.0147506 | 0.0609091 |
| GB41909 | bluestreak isoform X1 | 135.62904 | -0.3175723 | -0.1257869 | 0.1111989 | 0.0324056 |
| GB52352 | WD repeat and HMG-box DNA-binding protein 1-like isoform X1 | 135.59821 | -0.7548754 | 0.0713677 | -0.0431087 | 0.0860159 |
| GB42770 | male-specific lethal 3 homolog isoform X1 | 135.58846 | -0.5758687 | 0.2120173 | 0.0024509 | 0.2049724 |
| GB50801 | serine/threonine-protein kinase VRK1-like | 135.57948 | -0.1166064 | 0.0151375 | 0.0469310 | 0.1496590 |
| GB46615 | rho GTPase-activating protein 1-like isoformX2 | 135.55327 | -0.2261302 | 0.2031633 | 0.1223534 | -0.0680579 |
| GB46606 | peptidyl-prolyl cis-trans isomerase FKBP8-like isoform X3 | 135.41966 | -0.2334119 | -0.1754440 | -0.1659490 | 0.0951442 |
| GB50281 | transducin (beta)-like 3 isoform X1 | 135.39669 | -0.2836537 | 0.0159128 | 0.1175775 | 0.1492657 |
| GB45280 | mitochondrial import inner membrane translocase subunit TIM44-like isoform 1 | 135.33746 | -0.4501343 | 0.1405507 | -0.0166811 | 0.1794187 |
| GB41884 | calcineurin-binding protein cabin-1-like isoform X2 | 135.22346 | -0.1203653 | 0.1262933 | -0.1019124 | -0.1088786 |
| GB51658 | ribosome maturation protein SBDS-like | 135.22141 | 0.1290948 | 0.1091363 | 0.0419986 | 0.1668842 |
| GB42846 | cullin-4B-like | 135.21839 | -0.5256070 | 0.0567609 | 0.1313509 | -0.6419503 |
| GB51910 | zinc finger CCCH-type with G patch domain-containing protein-like | 135.17632 | -0.8185470 | 0.1433237 | 0.0578685 | 0.0006587 |
| GB53979 | SEC23-interacting protein-like isoformX1 | 135.05810 | -0.0502786 | 0.0270162 | 0.0947356 | -0.0260509 |
| GB42420 | coiled-coil domain-containing protein 102A-like | 135.04799 | -0.6434837 | 0.1403241 | 0.0143503 | 0.1137371 |
| 102655679 | DNA-directed RNA polymerase III subunit RPC7-like | 135.03891 | -0.0255525 | 0.2383947 | 0.1831898 | 0.0764651 |
| GB55281 | TBC1 domain family member 20-like isoform X2 | 134.96415 | 0.0672651 | -0.1426891 | 0.0804238 | 0.0657347 |
| GB47428 | synaptojanin-1 | 134.96298 | -0.1267762 | 0.2170341 | 0.1723928 | -0.0359031 |
| GB41469 | septin-2 isoform X1 | 134.75725 | 0.1015160 | -0.0625438 | 0.0466189 | 0.0969068 |
| GB43626 | uncharacterized protein LOC409331 isoform X3 | 134.68611 | 0.0623315 | 0.2804728 | -0.0700666 | -0.0088614 |
| GB54680 | LOW QUALITY PROTEIN: vinculin | 134.66846 | -0.2567412 | -0.1122626 | 0.1933519 | 0.1346479 |
| GB55854 | ubiquitin-protein ligase E3B-like isoform X3 | 134.59375 | -0.5010630 | -0.0673886 | 0.0766717 | -0.0285551 |
| GB49124 | myotubularin-related protein 2 isoform X3 | 134.53570 | 0.0859800 | 0.1215036 | -0.0566260 | -0.0406390 |
| GB46645 | ras-related protein Rab-21 | 134.38127 | -0.1477973 | -0.0186089 | 0.0715747 | -0.0108318 |
| GB53931 | laminin subunit alpha-1-like isoform X3 | 134.35243 | -0.4397656 | 0.0126489 | 0.7125136 | -0.6395672 |
| GB49344 | serine/threonine-protein kinase pelle isoform X1 | 134.23598 | -0.2198826 | -0.0143871 | 0.1806204 | 0.0907204 |
| GB52757 | uncharacterized protein LOC408473 isoform 1 | 134.22126 | -0.1246194 | -0.4210634 | 0.1312857 | 0.0290533 |
| GB45126 | DNA topoisomerase 1 isoform 2 | 134.18310 | -0.2462215 | 0.1326847 | -0.1616299 | 0.0858320 |
| GB49961 | WD repeat-containing protein 44-like isoform X1 | 134.07229 | -0.0883861 | -0.0441383 | 0.1789031 | 0.1614347 |
| 409254 | protein RTF2 homolog | 134.04799 | -0.2523550 | -0.2980123 | 0.0513461 | 0.2236932 |
| GB49520 | BRCA1-A complex subunit BRE-like | 134.03705 | -0.1608837 | 0.1074496 | 0.1093796 | 0.1509256 |
| GB44327 | protein SERAC1-like isoform X2 | 134.01956 | 0.0675840 | 0.0758677 | 0.1203074 | 0.0336178 |
| GB44429 | DNA damage-binding protein 1-like | 133.86472 | -0.0129277 | 0.1175625 | 0.0715284 | 0.0762634 |
| GB52421 | protein lines isoform 1 | 133.78321 | 0.0154913 | -0.0189455 | 0.0735825 | -0.0471184 |
| GB47594 | golgin-45-like | 133.76255 | -0.2228597 | 0.0565988 | -0.0394860 | -0.0497044 |
| GB41775 | integrator complex subunit 5-like isoform X2 | 133.67837 | -0.0170800 | 0.0127716 | -0.0699756 | -0.1695111 |
| GB52253 | protein PRRC2C-like isoform X2 | 133.58713 | -0.5419796 | -0.0766539 | 4.6580792 | 0.0782022 |
| GB53133 | ral GTPase-activating protein subunit beta-like isoform X7 | 133.42935 | -0.1036454 | -0.0753829 | 0.1748863 | -0.2134538 |
| GB46654 | ribosomal L1 domain-containing protein CG13096-like | 133.30715 | -0.2395835 | -0.0102828 | -0.0263839 | 0.0368059 |
| GB55591 | microtubule-associated protein futsch-like isoform X1 | 133.30691 | -0.4632335 | -0.2611452 | -1.1496085 | -0.3570381 |
| GB49315 | protein lin-54 homolog isoform X2 | 133.24962 | -0.2850337 | 0.1621247 | 0.0445596 | 0.4659218 |
| GB51597 | DNA repair protein complementing XP-A cells homolog | 133.23296 | -0.2223217 | 0.0855791 | -0.1372374 | 0.0965293 |
| GB47436 | acetyl-CoA acetyltransferase, mitochondrial-like isoform X1 | 133.18470 | 0.0417805 | -0.0757904 | 0.0249588 | -0.1616918 |
| GB49033 | glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial isoform X1 | 133.07916 | 0.2616057 | -0.0117697 | -0.0320500 | -0.0887660 |
| GB53439 | putative sodium-coupled neutral amino acid transporter 10-like isoform X2 | 133.06697 | -0.6416222 | -0.0673806 | -0.0277337 | 0.6700455 |
| GB46881 | COMM domain-containing protein 5-like isoform X2 | 133.04645 | -0.1888059 | 0.2031945 | -0.0252253 | 0.1646374 |
| GB47238 | UPF0396 protein CG6066-like isoform 1 | 133.02286 | -0.1716189 | 0.1377160 | -0.0236372 | 0.1211281 |
| GB50269 | very-long-chain (3R)-3-hydroxyacyl-[acyl-carrier protein] dehydratase 3 isoform X2 | 132.94712 | -0.3144725 | 0.1069885 | 0.1133108 | 0.0413660 |
| GB46627 | paraplegin-like | 132.92270 | -0.6454774 | -0.0089053 | 0.0224408 | 0.1257638 |
| GB41640 | zucchini | 132.80851 | -0.0803640 | 0.1773846 | -0.0533377 | 0.0770383 |
| GB46691 | histone-lysine N-methyltransferase, H3 lysine-79 specific-like isoform X2 | 132.77865 | -0.1398413 | 0.1288700 | 0.0697022 | -0.0789529 |
| GB53707 | density-regulated protein-like isoform X1 | 132.61700 | -0.0526849 | 0.0986741 | -0.0239636 | 0.0103761 |
| GB40634 | upstream stimulatory factor 1-like | 132.53514 | -0.0683351 | 0.0005669 | 0.1678361 | 0.1358999 |
| GB45279 | RING finger protein PFF0165c-like | 132.38270 | -0.5602977 | 0.0443648 | -0.0806039 | 0.1729593 |
| GB46216 | protein RFT1 homolog isoform X1 | 132.31517 | -0.0937464 | -0.0258783 | -0.0282630 | 0.0568698 |
| GB48370 | ATP-binding cassette sub-family B member 7, mitochondrial isoform X1 | 132.29126 | -0.5912863 | -0.1632948 | 0.0587712 | 0.1400553 |
| GB53960 | spliceosome-associated protein CWC15 homolog isoform 1 | 132.27536 | 0.2116932 | 0.1324830 | -0.0340064 | -0.0275712 |
| GB49680 | LIM domain kinase 1 isoform X2 | 132.27187 | -0.0530115 | 0.1548388 | 0.1821953 | 0.1045718 |
| GB53882 | IST1 homolog isoform X4 | 132.10557 | -0.2744962 | -0.0095574 | -0.0028003 | 0.1880862 |
| GB49762 | nuclear pore complex protein Nup50 isoform X1 | 131.99821 | -0.2926662 | 0.0635457 | -0.0599136 | -0.0029573 |
| GB55439 | probable ATP-dependent RNA helicase DDX49-like | 131.92893 | -0.5517570 | -0.0107095 | 0.1729551 | 0.0900870 |
| GB53068 | cysteine and histidine-rich domain-containing protein | 131.78883 | 0.0529541 | 0.1320168 | 0.1017358 | 0.1695318 |
| GB53618 | uncharacterized protein LOC724843 isoform X1 | 131.71954 | -0.7218422 | -0.1571234 | 0.0176053 | -0.0791466 |
| GB55183 | ankyrin repeat domain-containing protein SOWAHB-like isoform X5 | 131.49345 | -0.7243208 | 0.0317372 | 0.2646035 | -0.2066738 |
| GB54672 | 39S ribosomal protein L21, mitochondrial-like | 131.49019 | 0.0141550 | 0.1263036 | -0.0528228 | 0.2099445 |
| GB47596 | general transcription factor 3C polypeptide 1-like | 131.45492 | -0.6789068 | -0.0444361 | 0.1979938 | -0.0472513 |
| GB40909 | sorting nexin-29-like isoform X4 | 131.45347 | -0.1331307 | -0.2158390 | 0.1620283 | -0.0533020 |
| GB49032 | CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase, mitochondrial-like isoform X2 | 131.39431 | 0.4081832 | -0.1031490 | -0.0136587 | 0.0103266 |
| GB47576 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 5-like isoform 2 | 131.38395 | -0.1467968 | -0.0623679 | -0.1063544 | 0.2049577 |
| GB44062 | tyrosine-protein kinase Fps85D-like isoform X4 | 131.34635 | -0.0953993 | -0.0522888 | 0.1602647 | 0.1262338 |
| GB52645 | pinin | 130.95168 | -0.4535884 | 0.1161390 | 0.1413056 | 0.0958147 |
| GB55626 | peptidyl-prolyl cis-trans isomerase D-like isoform X1 | 130.90875 | -0.3070889 | 0.1112651 | -0.0078134 | 0.0884890 |
| GB46257 | myotubularin-related protein 14 isoform 1 | 130.82134 | -0.0748272 | 0.2625850 | 0.1223574 | 0.1265138 |
| GB45457 | vacuolar protein sorting-associated protein 53 homolog | 130.76178 | -0.2682224 | 0.1298956 | 0.1668593 | -0.0116516 |
| GB53939 | mitotic checkpoint protein BUB3 | 130.52660 | -0.1194331 | 0.1200242 | -0.0198973 | 0.0907735 |
| GB54592 | tudor domain-containing protein 7-like isoform X1 | 130.49148 | 0.0634736 | 0.0356082 | 0.1425800 | 0.1258433 |
| GB46736 | probable ubiquitin carboxyl-terminal hydrolase FAF-X isoform X6 | 130.42037 | -0.4474171 | 0.1103373 | -0.8116499 | -0.0098906 |
| GB41770 | 28S ribosomal protein S22, mitochondrial | 130.37631 | 0.1971519 | 0.1247985 | 0.0801954 | -0.0128793 |
| GB51134 | serine/threonine-protein kinase mig-15 isoform X13 | 130.26925 | -0.2997902 | -0.0414082 | 0.1044629 | -0.0001478 |
| GB46048 | transmembrane protein 62-like, transcript variant X2 | 130.19641 | -0.5460422 | 0.0065798 | 0.0505555 | 0.4044524 |
| GB47991 | biogenesis of lysosome-related organelles complex 1 subunit 1-like | 130.14915 | 0.0521281 | 0.0402486 | 0.1343298 | 0.0696550 |
| GB53820 | gametogenetin-binding protein 2-like isoform X1 | 130.14014 | 0.0355411 | -0.0413955 | -0.0791741 | 0.1307555 |
| GB51395 | rabenosyn-5 | 130.10966 | -0.1867821 | 0.1416788 | 0.0541704 | 0.0742042 |
| GB45545 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9-like | 130.03078 | -0.3035913 | -0.1743814 | 0.0395625 | 0.1680090 |
| 102655877 | beta-1,3-galactosyltransferase 6-like | 129.92442 | -0.3488118 | 0.0705781 | 0.2429741 | 0.0330696 |
| GB50897 | glycoprotein 150 isoform X1 | 129.77615 | -0.2437159 | -0.1423997 | 0.0155776 | -0.0290678 |
| GB51542 | PH-interacting protein isoform X1 | 129.75422 | -0.1598458 | -0.1386128 | 0.1681110 | 0.1289269 |
| GB46740 | N-acetylgalactosaminyltransferase 6-like isoform 1 | 129.69168 | -0.2175312 | 0.0044725 | 0.0234961 | -0.0840065 |
| GB43873 | sin3 histone deacetylase corepressor complex component SDS3-like isoform 1 | 129.58139 | 0.1197990 | 0.1093941 | -0.0390499 | 0.2543805 |
| GB47747 | 60S ribosomal protein L6 | 129.55702 | -0.0038808 | -0.0176271 | 0.1970132 | 0.1608325 |
| GB55492 | importin-9 isoform X2 | 129.23748 | 0.1007102 | 0.1599284 | -0.2256385 | 0.0205695 |
| GB42721 | aspartate–tRNA ligase, mitochondrial-like | 129.19710 | -0.0632949 | -0.0089592 | 0.2995115 | 0.1006103 |
| GB50764 | coiled-coil domain-containing protein 124-like | 129.11705 | -0.1298397 | 0.1096236 | -0.1791570 | 0.1126657 |
| GB51067 | akirin-2 isoform 2 | 128.88890 | 0.3690387 | 0.1098612 | -0.0267994 | -0.1043719 |
| GB44890 | regulator of nonsense transcripts 1 isoform X1 | 128.82290 | -0.2037780 | 0.0141096 | 0.0179000 | -0.1040571 |
| GB49402 | erlin-1-like | 128.71228 | -0.3886355 | 0.1079698 | 0.0151644 | 0.2204134 |
| GB51884 | calcyclin-binding protein-like | 128.46540 | -0.0633120 | 0.1493639 | 0.0745391 | 0.0277878 |
| GB45342 | transcriptional adapter 1-like isoform 1 | 128.45373 | -0.2026429 | 0.1628707 | -0.0169983 | 0.0487770 |
| GB41304 | probable glutamine–tRNA ligase | 128.42973 | -0.0782771 | -0.0067491 | -0.1096354 | -0.1321491 |
| GB47846 | WD repeat-containing protein 48-like isoform X2 | 128.26954 | 0.1051237 | 0.0446909 | 0.0761726 | 0.0944745 |
| GB41693 | DNA topoisomerase 3-alpha-like | 127.95605 | -0.0649003 | 0.0211336 | -0.0604116 | 0.1850720 |
| GB43868 | dnaJ homolog subfamily C member 1-like isoform X1 | 127.94984 | -0.3941120 | 0.1522078 | -0.0622720 | -0.1150482 |
| GB40091 | LIM domain-containing protein jub-like | 127.85360 | 0.0708016 | 0.0090090 | 0.0481691 | 0.0941938 |
| GB47960 | moesin/ezrin/radixin homolog 1 isoform X15 | 127.80969 | 0.1435555 | -0.0593738 | 0.0479489 | 0.0214360 |
| GB49354 | uncharacterized protein PF11_0213-like isoform X1 | 127.78012 | -0.2457494 | 0.2333305 | -0.0192781 | 0.1512150 |
| GB42071 | GATA zinc finger domain-containing protein 7-like | 127.75657 | -0.2207894 | -0.0076410 | 0.0248166 | 0.0459783 |
| GB46378 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase I | 127.69226 | 0.2109927 | -0.0067827 | 0.8315764 | 0.0480976 |
| GB55422 | maspardin-like isoform X3 | 127.63431 | 0.0029986 | 0.1376321 | 0.1030255 | 0.0651460 |
| GB51427 | bromodomain-containing protein DDB_G0280777 | 127.63373 | -0.4958885 | 0.2153698 | -0.1018241 | 0.1583879 |
| GB55570 | transmembrane protein 53-like isoform X3 | 127.58972 | -0.3618078 | -0.1642811 | 0.4374619 | 0.1062865 |
| GB43260 | macrophage erythroblast attacher-like isoform 1 | 127.36079 | 0.0446683 | 0.1894613 | 0.0317748 | 0.0349377 |
| GB40098 | cyclin-H | 127.30857 | -0.0418528 | 0.1194292 | 0.5277859 | 0.2339003 |
| GB45037 | beta-lactamase-like protein 2-like isoform X2 | 127.28879 | -0.5724075 | 0.0028505 | -0.3561338 | -0.4453576 |
| 102654890 | activating signal cointegrator 1-like isoform X2 | 127.19822 | -0.2278917 | 0.0238683 | -0.1951713 | 0.1136874 |
| GB40491 | translation initiation factor eIF-2B subunit gamma isoform X1 | 127.15652 | -0.0249244 | -0.0772291 | 0.0772539 | 0.0629394 |
| GB44069 | syntaxin-16 isoform X1 | 127.06668 | -0.7860325 | 0.2135015 | 0.4228089 | 0.1496153 |
| 102656215 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like | 127.03149 | -0.6157903 | 0.1121564 | 0.1025515 | 0.0690541 |
| GB50515 | ankycorbin-like isoform X1 | 126.98409 | -0.4791568 | 0.0637900 | 0.1688905 | 0.0063271 |
| GB43301 | ras GTPase-activating protein-binding protein 2 isoform X1 | 126.96412 | 0.0577429 | 0.0105586 | 0.0778471 | 0.1607821 |
| GB49500 | zinc finger protein 808-like, transcript variant X2 | 126.93821 | -0.6101884 | 0.1800285 | -0.0489694 | 0.0743351 |
| GB53184 | replication protein A 32 kDa subunit | 126.82564 | -0.0395673 | 0.1803790 | 0.1233143 | 0.0737084 |
| GB44660 | probable ribonuclease P/MRP protein subunit POP5-like isoform 2 | 126.74854 | -0.1652242 | 0.0253384 | 0.0433548 | 0.0390910 |
| GB52658 | general transcription factor 3C polypeptide 5-like | 126.56580 | -0.3274466 | 0.0989520 | 0.1706715 | 0.0243821 |
| GB51763 | UPF0505 protein C16orf62 homolog isoform X1 | 126.45012 | -0.1913434 | -0.0666195 | 0.1090036 | -0.0315871 |
| GB52994 | transcription elongation factor B polypeptide 1 isoform X5 | 126.42032 | -0.0128444 | 0.1146241 | 0.1496385 | 0.1702401 |
| GB46268 | plasminogen activator inhibitor 1 RNA-binding protein-like isoform X1 | 126.39903 | 0.2156731 | 0.1094758 | -0.0154047 | -0.0077640 |
| GB46042 | TBC1 domain family member 23-like | 126.38852 | 0.1655253 | 0.0469410 | -0.0151648 | 0.1450667 |
| GB47271 | adenylate cyclase type 9-like isoform X2 | 126.32175 | -0.1380968 | -0.0195978 | -0.0258853 | 0.0999105 |
| GB53836 | protein spinster-like isoformX1 | 126.24559 | 0.0453729 | -0.1501046 | 0.0064097 | 0.0151537 |
| GB40467 | ubiquitin-conjugating enzyme E2 J2-like | 126.18787 | -0.0201480 | 0.0753651 | -0.0657088 | 0.0230615 |
| 726689 | prefoldin subunit 1-like isoform X1 | 126.17229 | 0.0610065 | -0.0389106 | 0.0012349 | 0.2877932 |
| GB47539 | acidic mammalian chitinase isoform X2 | 125.99563 | 0.1265903 | 0.0132536 | 0.2027685 | 0.1587172 |
| GB40872 | josephin-like protein-like isoform X1 | 125.85313 | -0.4059615 | 0.0073056 | 0.0371880 | 0.3698352 |
| GB54340 | GTP-binding protein 10 homolog | 125.80689 | -0.1804535 | 0.1012403 | 0.0558442 | 0.0854720 |
| GB54928 | tRNA (guanine(10)-N2)-methyltransferase homolog | 125.78091 | -0.1690325 | 0.0448066 | -0.0121400 | 0.2709146 |
| GB46915 | serine/threonine-protein phosphatase 2A activator isoform X2 | 125.74771 | 0.0758897 | 0.0835348 | 0.1491264 | 0.0811278 |
| 725981 | protein TIPIN homolog | 125.74518 | 0.0640545 | -0.0477598 | -0.0318140 | 0.1070849 |
| GB48417 | probable E3 ubiquitin-protein ligase RNF144A-like isoform X1 | 125.69744 | -0.3765713 | -0.0200665 | 0.0460438 | 0.1067988 |
| GB53196 | uncharacterized protein LOC412107 | 125.67399 | -0.3512672 | -0.1425177 | -0.0109280 | 0.1654680 |
| GB48351 | succinate-semialdehyde dehydrogenase, mitochondrial | 125.53193 | 0.2143112 | 0.0627905 | -0.0843891 | -0.0230087 |
| GB51037 | CTD small phosphatase-like protein 2-like isoform X1 | 125.44515 | 0.0195500 | -0.0410946 | -0.0197439 | 0.1016195 |
| GB49221 | phosphomevalonate kinase-like | 125.42336 | 0.5112757 | -0.2234616 | -0.1354606 | 0.0078624 |
| GB53663 | GTPase Era, mitochondrial-like isoform X1 | 125.41223 | -0.4083925 | -0.0622608 | 0.0927247 | 0.0529931 |
| GB52918 | serine protease HTRA2, mitochondrial | 125.25263 | 0.0268476 | 0.0311616 | -0.0062193 | -0.0593778 |
| GB44144 | probable phenylalanine–tRNA ligase, mitochondrial | 125.23986 | -0.7019202 | 0.0469811 | 0.1340443 | 0.2442396 |
| GB47272 | nucleoporin Nup37-like isoform X2 | 125.21865 | 0.4564698 | 0.4591604 | -0.1252003 | 0.1746374 |
| GB51721 | zinc finger protein 800-like isoform X2 | 125.14502 | -0.8962219 | 0.1574114 | 0.1203346 | 0.1977810 |
| GB47746 | AP-3 complex subunit beta-2-like isoform X3 | 125.13494 | -0.3338172 | 0.1004985 | -0.0023056 | 0.0984828 |
| GB47577 | SAP domain-containing ribonucleoprotein-like isoform X3 | 124.96337 | -0.0891455 | 0.2085310 | -0.0489876 | 0.1285256 |
| GB48122 | MAU2 chromatid cohesion factor homolog isoform X1 | 124.95705 | -0.2028627 | 0.0556739 | 0.0333472 | 0.0353484 |
| GB44830 | cleavage stimulation factor subunit 2 isoform X2 | 124.92111 | 0.2849909 | 0.1727879 | 0.1058351 | -0.1563559 |
| GB40017 | enhancer of mRNA-decapping protein 4 | 124.83483 | -0.1405951 | 0.1520184 | -0.1054822 | 0.0233113 |
| GB46274 | arginine/serine-rich coiled-coil protein 2-like isoform X2 | 124.65000 | -0.3269109 | -0.0600284 | 0.1876375 | -0.2644653 |
| GB49777 | seipin-like isoform 1 | 124.64821 | 0.0130656 | 0.0234229 | -0.0599748 | 0.1408062 |
| GB55506 | histidine–tRNA ligase, cytoplasmic-like isoform X5 | 124.44594 | 0.1707464 | 0.0409954 | 0.0624129 | 0.0210656 |
| GB49556 | DNA replication licensing factor Mcm2-like isoform X1 | 124.40342 | -0.6415868 | 0.0286972 | 0.0286895 | 0.0290473 |
| GB42786 | microtubule-associated protein RP/EB family member 1-like isoform X4 | 124.38501 | 0.3721071 | 0.1083819 | 0.0799526 | 0.1225451 |
| GB54354 | uncharacterized protein DDB_G0274915-like isoform X2 | 124.30810 | 0.1084176 | 0.0363753 | -0.5016934 | 0.0447914 |
| GB54976 | ER membrane protein complex subunit 8/9 homolog | 124.29445 | 0.1352807 | -0.0593081 | -0.0122129 | 0.0506201 |
| GB47007 | actin-related protein 5 | 124.26723 | -0.0997846 | -0.0481023 | 0.0215667 | 0.2181803 |
| GB41252 | cell cycle checkpoint protein RAD17-like isoform X2 | 123.97373 | 0.0958116 | 0.0581361 | -0.0410770 | 0.0397078 |
| GB53677 | clavesin-2-like isoform X3 | 123.87925 | -0.5582742 | 0.1597406 | 0.1839171 | -0.0989334 |
| 100577180 | uncharacterized protein LOC100577180 | 123.80272 | 0.0091102 | 0.1765232 | 0.0391926 | 0.1001481 |
| GB53659 | ribosome biogenesis protein WDR12 homolog isoform X1 | 123.79581 | 0.1977254 | 0.2142607 | -1.3574753 | 0.0468377 |
| GB48223 | peroxisomal targeting signal 1 receptor-like isoform X1 | 123.77770 | -0.1573646 | -0.0057346 | -0.0039700 | 0.0379803 |
| GB55003 | protein phosphatase 1 regulatory subunit 16A-like isoform X7 | 123.77505 | -0.1071349 | 0.0732572 | 0.1043548 | 0.1321670 |
| GB50727 | prefoldin subunit 2-like | 123.68054 | 0.2567572 | 0.0130137 | 0.0759218 | 0.0508393 |
| GB44904 | serine/threonine-protein phosphatase 4 regulatory subunit 2 | 123.61833 | -0.9442707 | 0.0768056 | -0.0239977 | -0.0365701 |
| GB46751 | nuclear export mediator factor NEMF homolog isoform X2 | 123.57726 | 0.0005700 | 0.0950236 | 0.0404740 | -0.0290170 |
| GB45272 | neuroligin-4, Y-linked isoform X2 | 123.57192 | -0.5009508 | 0.0028581 | -0.0385243 | -0.0257233 |
| GB46204 | protein cereblon-like | 123.52119 | 0.1390750 | -0.0760547 | -0.0126805 | 0.1431147 |
| GB41850 | hemocytin isoform X3 | 123.44266 | -0.0058567 | 0.0205642 | -0.4043645 | 0.2052439 |
| GB45716 | WD repeat and FYVE domain-containing protein 2-like | 123.29325 | -0.4528887 | -0.1209118 | 0.0596195 | 0.1341635 |
| GB54287 | protein virilizer | 123.22607 | -0.0221346 | 0.0681456 | 0.0054647 | -0.0620630 |
| GB41213 | intraflagellar transport protein 20 homolog isoform X1 | 123.18194 | -0.1197624 | 0.0062110 | 0.0078017 | -0.0140481 |
| GB40955 | glutamate–cysteine ligase regulatory subunit isoform X2 | 123.09107 | 0.1282528 | -0.0324026 | -0.0222198 | -0.0866865 |
| 102654121 | protein YIPF5-like | 122.98313 | 0.2537020 | 0.1358925 | 0.0051856 | -0.1262311 |
| GB49593 | cytosolic carboxypeptidase-like protein 5-like isoform X1 | 122.94352 | -0.5903926 | -0.2785982 | 0.1007198 | 0.2362319 |
| GB51087 | pyridoxal kinase-like | 122.91547 | -0.5788949 | -0.0323483 | 0.0467033 | 0.0958125 |
| GB53726 | protein CNPPD1-like | 122.88417 | -0.0813809 | 0.1552521 | 0.0434168 | 0.1190650 |
| GB54579 | regulator of nonsense transcripts 3B isoform 1 | 122.84881 | -0.5184260 | 0.0407833 | 0.1826402 | -0.0038804 |
| GB51158 | uncharacterized protein LOC726321 | 122.82610 | -0.1685591 | -0.0550626 | 0.0757449 | 0.1664207 |
| GB47633 | serine/threonine-protein kinase D3 isoformX2 | 122.80513 | -0.4167619 | 0.0384036 | 0.0536617 | 0.0302385 |
| GB51508 | probable elongator complex protein 3-like isoform X3 | 122.74449 | 0.0860960 | 0.2219367 | 0.7217703 | 0.0817006 |
| GB48204 | CD109 antigen isoform X2 | 122.70403 | -0.1895645 | -0.0868434 | -0.0159124 | -0.1811137 |
| GB44906 | ubiquitin conjugation factor E4 B isoform X1 | 122.69198 | -0.3744256 | -0.0109923 | -0.1292852 | 0.0396984 |
| GB49538 | inhibitor of growth protein 1-like | 122.58239 | -0.5570093 | 0.2187936 | -0.0883342 | 0.0977171 |
| GB45568 | uncharacterized protein C2orf42 homolog | 122.28137 | -0.6094268 | 0.1540253 | 0.0750140 | 0.1139424 |
| GB42079 | protein arginine N-methyltransferase 8 | 122.24493 | 0.5733701 | -0.0329699 | 0.0139856 | -0.0079264 |
| GB48010 | protein ENL-like isoform X2 | 122.19029 | -0.1762306 | 0.1021199 | 0.0919987 | -0.0182488 |
| GB47844 | AP-3 complex subunit delta-1 | 121.98324 | -0.4769263 | -0.0609007 | -0.0141209 | 0.0865234 |
| GB42778 | U6 snRNA-associated Sm-like protein LSm4-like | 121.95927 | 0.2395203 | 0.1258932 | 0.0384109 | -0.0247357 |
| GB43622 | endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase-like isoform X1 | 121.93125 | -0.2287433 | -0.0176362 | -0.0707171 | 0.1184653 |
| GB41969 | mediator of RNA polymerase II transcription subunit 26 isoform X3 | 121.89217 | -0.2091069 | 0.1411050 | 0.2147840 | -0.0488400 |
| 725795 | INO80 complex subunit D-like | 121.87241 | 0.0762136 | -0.0682986 | 0.0363835 | 0.1172964 |
| GB55012 | transcription factor Dp-1 | 121.84800 | 0.0615153 | 0.2397292 | 0.1097454 | 0.9006222 |
| GB47554 | apoptosis inhibitor 5-like | 121.69504 | -0.1868560 | 0.0642729 | 0.1980508 | 0.0854192 |
| GB54968 | phosphorylated adapter RNA export protein | 121.57861 | -0.1946227 | 0.0675115 | 0.1038418 | 0.1358846 |
| GB46962 | SH3 domain-binding glutamic acid-rich protein homolog isoform X1 | 121.52820 | 0.2925105 | 0.0673425 | 0.1769261 | 0.1212187 |
| GB46773 | cysteine-rich protein 2-binding protein-like | 121.39440 | -0.4039526 | 0.1121328 | 0.0121977 | 0.1249059 |
| GB51589 | nuclear pore membrane glycoprotein 210-like | 121.35661 | 0.4097703 | 0.0470867 | -0.4446937 | -0.0472264 |
| GB53717 | uncharacterized protein LOC726365 isoform X2 | 121.20585 | -0.4442012 | 0.0587499 | 0.1435022 | -0.0778604 |
| GB50032 | prefoldin subunit 6-like isoform X1 | 121.07031 | 0.2472118 | -0.2161193 | 0.0589943 | 0.1217440 |
| GB41754 | protein DDI1 homolog 2-like isoform X2 | 121.01607 | 0.0928256 | -0.0753114 | 0.0186832 | 0.1555542 |
| 102655440 | uncharacterized protein LOC102655440 | 120.99542 | -0.6491477 | 0.0444111 | 0.1100712 | 0.1183861 |
| GB54019 | myosin-1-like isoform X2 | 120.96709 | -0.0241462 | 0.1489033 | 0.1169968 | 0.0254983 |
| GB49883 | uncharacterized protein CG7065-like | 120.63533 | -0.0001075 | -0.0408664 | 0.0346524 | 0.0401417 |
| GB44049 | spermatogenesis-associated protein 5-like isoform X1 | 120.59942 | 0.0870816 | -0.0981649 | 0.1035189 | 0.1074495 |
| GB47427 | protein ariadne-2-like | 120.57182 | -0.0655958 | 0.0650006 | -0.0509156 | 0.0374485 |
| 551815 | uncharacterized protein C9orf114 homolog | 120.33450 | 0.1003637 | 0.1337426 | 0.0943394 | -0.2370965 |
| GB41455 | cysteine–tRNA ligase, cytoplasmic-like | 120.33315 | 0.0392605 | -0.0313631 | 0.0524425 | 0.2351291 |
| GB52801 | phosphatidylinositol 5-phosphate 4-kinase type-2 beta-like isoformX1 | 120.31313 | -0.1409585 | -0.1214348 | 0.2551378 | 0.0593211 |
| GB50555 | thioredoxin peroxidase 3 isoform 2 | 120.29327 | 0.1611813 | 0.0486416 | 0.6320661 | 0.0798716 |
| GB45107 | peroxisome biogenesis factor 1-like isoform X1 | 120.23905 | 0.1556906 | 0.2945277 | 0.0638324 | -0.0707098 |
| GB42751 | LOW QUALITY PROTEIN: DNA repair protein RAD50 | 120.16956 | -0.0433836 | -0.1596184 | 0.1787985 | 0.1607935 |
| GB48467 | methyltransferase-like protein 13-like isoform X2 | 120.13408 | -0.1119520 | 0.0574958 | 0.2750865 | -0.0083499 |
| GB42744 | uncharacterized protein LOC100577988 | 120.09494 | -0.2652405 | 0.0042741 | 0.4681334 | 0.0599145 |
| GB48247 | uncharacterized protein LOC410423 isoform X1 | 119.81933 | -0.3371425 | 0.2051331 | 0.0014905 | -0.0374518 |
| GB41140 | UPF0609 protein CG1218-like isoform X1 | 119.80031 | -0.7523746 | 0.1118520 | 0.3406966 | 0.1388311 |
| GB45709 | broad-complex core protein isoforms 1/2/3/4/5 isoform X6 | 119.68996 | -0.2249650 | -0.0039711 | 0.4763229 | 0.0518593 |
| GB53250 | PHD finger protein 12-like isoform X1 | 119.64041 | -0.3430417 | 0.0713771 | 0.0079793 | 0.0835927 |
| GB54385 | cation transport regulator-like protein 2-like | 119.55672 | -0.2764940 | -0.0538840 | 0.0536482 | -0.2872535 |
| GB53832 | lipoyltransferase 1, mitochondrial-like isoformX2 | 119.49156 | -0.2308916 | -0.2117358 | -0.0814708 | -0.0068559 |
| GB45695 | protein cramped-like isoform X2 | 119.44908 | -0.2846885 | -0.1967639 | 0.0640146 | 0.1065411 |
| GB41002 | protein timeless homolog isoform X3 | 119.40888 | -0.1157529 | 0.2487499 | 0.0645132 | -0.1332480 |
| GB45837 | transcription factor 25-like isoform X2 | 119.39937 | -0.4091535 | 0.0403150 | 0.0223616 | 0.1047016 |
| GB53398 | zinc finger protein 622-like isoform 1 | 119.29143 | -0.0217688 | -0.0061659 | -0.0588995 | 0.0773505 |
| GB42014 | E3 ubiquitin-protein ligase TRIP12-like isoform X3 | 119.17333 | -0.0545045 | -0.0142915 | 0.1006065 | 0.0041780 |
| GB45014 | nuclear pore complex protein Nup93-like | 119.09256 | 0.1809858 | 0.2185313 | 0.0794949 | -0.1472499 |
| GB54297 | programmed cell death protein 2 | 119.06425 | -0.8893836 | 0.0180097 | 0.3057900 | 0.1112449 |
| GB41489 | nuclear fragile X mental retardation-interacting protein 1-like isoform X1 | 119.04696 | 0.0155358 | 0.1158237 | 0.0491491 | 0.0189348 |
| 724372 | nucleolar protein 12-like isoform X1 | 119.04547 | -0.9134259 | 0.0959727 | -0.2595753 | -0.0694779 |
| GB54209 | prostatic acid phosphatase-like isoform X1 | 119.01385 | -0.6946473 | 0.1459312 | 0.1345460 | -0.4049664 |
| GB42200 | uncharacterized protein LOC408577 | 118.92969 | -0.0516659 | -0.0767156 | -0.1588003 | -0.3674519 |
| GB43213 | muskelin | 118.83908 | -0.1627108 | 0.1859857 | -0.0895491 | 0.0845601 |
| GB50704 | spindle assembly abnormal protein 6 homolog | 118.83559 | 0.0495763 | -0.0187462 | 0.0070448 | 0.0309649 |
| GB53538 | nischarin | 118.78685 | 0.1268951 | -0.1976658 | 0.1935602 | 0.0502124 |
| GB45507 | cell division cycle protein 23 homolog | 118.74905 | -0.2385612 | -0.0554456 | -0.0119803 | 0.0216882 |
| GB48789 | disks large homolog 5-like isoform X2 | 118.66292 | -1.3197825 | -0.0131642 | -0.0829889 | 0.0168027 |
| GB46194 | uncharacterized protein LOC725889 | 118.51732 | -0.1058467 | 0.1584688 | 0.6893503 | 0.0837315 |
| GB47827 | exosome complex component RRP45-like isoform X2 | 118.50282 | -0.3997369 | 0.1627799 | 0.0316627 | 0.1904206 |
| GB54530 | dedicator of cytokinesis protein 7-like | 118.39830 | -0.1409387 | -0.2570711 | 0.5958964 | 0.1077095 |
| GB41603 | PTB domain-containing adapter protein ced-6 isoform X2 | 118.27967 | -0.2280360 | 0.0148098 | -0.4736116 | -0.1136950 |
| 102656134 | transcriptional adapter 2B-like isoform X2 | 118.21934 | 0.0923514 | -0.0135443 | 0.5085743 | 0.0579195 |
| GB54433 | 60S ribosomal protein L19 | 118.12611 | 0.3380498 | 0.0439157 | -0.0423034 | -0.0053281 |
| 411879 | WD repeat-containing protein 82-like | 118.10677 | 0.2745613 | -0.0002336 | 0.1394681 | 0.1121620 |
| GB47016 | LOW QUALITY PROTEIN: kynurenine–oxoglutarate transaminase 3-like | 118.10615 | 0.1376502 | -0.1319999 | -0.0056947 | 0.0202601 |
| GB42714 | splicing factor 45-like | 118.02329 | 0.1632569 | 0.0940064 | -0.0558715 | -0.0012813 |
| GB49155 | lysine–tRNA ligase isoform X1 | 118.01783 | 0.1954440 | 0.0014015 | -0.0435963 | 0.0016166 |
| GB47965 | MKI67 FHA domain-interacting nucleolar phosphoprotein-like | 117.97305 | -0.2029845 | 0.0070633 | 0.0427264 | 0.0366525 |
| GB53199 | N-acetylgalactosaminyltransferase 7 | 117.87922 | 0.0379632 | 0.1280538 | -0.0674433 | -0.0038215 |
| GB40886 | CCA tRNA nucleotidyltransferase 1, mitochondrial-like isoform X2 | 117.78899 | -0.7085378 | -0.0262270 | -0.0970282 | -0.0306402 |
| GB45666 | protein arginine N-methyltransferase 3 | 117.73896 | -0.4486753 | 0.1301691 | -0.2496239 | -0.0423792 |
| GB47299 | bile salt-activated lipase-like isoform X4 | 117.71662 | 0.1712936 | -0.2264894 | -0.1647928 | -0.1649715 |
| GB49828 | zinc finger CCHC-type and RNA-binding motif-containing protein 1-like | 117.60240 | -0.1924582 | 0.0943031 | 0.0809559 | -0.1410056 |
| GB48857 | quinone oxidoreductase-like protein 2-like | 117.59524 | 0.3740938 | 0.0973693 | 0.1411908 | 0.0891799 |
| GB47589 | cell division cycle and apoptosis regulator protein 1 | 117.48402 | -0.6034875 | 0.0571287 | -0.0059509 | 0.1193053 |
| GB53223 | rhomboid-related protein 3-like isoform X1 | 117.47946 | 0.0970346 | -0.1594247 | 0.0424554 | 0.0386046 |
| GB50130 | nucleolar complex protein 3 homolog | 117.46991 | -0.1607314 | 0.2280594 | 0.0982170 | 0.1048403 |
| GB49457 | hydroxymethylglutaryl-CoA lyase, mitochondrial-like isoform X1 | 117.46715 | -0.2959583 | 0.0575392 | -0.0105712 | -0.1225494 |
| GB54049 | eukaryotic translation initiation factor 3 subunit L-like | 117.46284 | 0.1002412 | 0.0566860 | -0.0162669 | -0.1057540 |
| GB55804 | kelch domain-containing protein 10 homolog isoform 2 | 117.32005 | 0.2269868 | -0.0937568 | 0.1937740 | 0.1023185 |
| GB44140 | WW domain-binding protein 4-like | 117.31191 | 0.0271393 | -0.0313370 | 0.1012644 | -0.0578627 |
| GB46889 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1-like | 117.02973 | -0.7178584 | -0.0050299 | 0.1329447 | 0.2546890 |
| GB48682 | protein FAM50 homolog | 117.00368 | 0.0589199 | 0.2917228 | 0.1116129 | 0.3004882 |
| GB48103 | syntaxin-12 isoform X1 | 116.98129 | -0.1808274 | -0.0425025 | -0.0163089 | 0.1177951 |
| 102656501 | putative uncharacterized zinc finger protein 814-like | 116.97038 | 0.0824778 | -0.0777484 | 0.1060117 | -0.0332942 |
| GB41231 | ubiquitin-conjugating enzyme E2 G1 | 116.93866 | 0.1151667 | 0.1193910 | -0.0606040 | 0.0372116 |
| GB45449 | protein LZIC-like | 116.91208 | -0.0162891 | -0.0106699 | -0.1697473 | 0.1709688 |
| GB51505 | sphingomyelin phosphodiesterase 4-like | 116.89204 | 0.1305566 | 0.0337235 | 0.0787225 | 0.1182446 |
| GB55517 | uncharacterized protein LOC410000 isoform X1 | 116.84710 | -0.3566292 | 0.0440213 | -0.6104877 | 0.1254497 |
| GB55477 | 39S ribosomal protein L19, mitochondrial | 116.84411 | -0.3137930 | 0.1760760 | -0.8871225 | 0.0961409 |
| GB44758 | eukaryotic translation initiation factor 2-alpha kinase isoform X3 | 116.58269 | -0.2238442 | -0.0861365 | -0.1326210 | 0.2470978 |
| GB46898 | transcription initiation factor TFIID subunit 7 isoform X3 | 116.52887 | -0.0169386 | 0.1229395 | 0.0364866 | 0.1798703 |
| GB44082 | engulfment and cell motility protein 1 | 116.52437 | 0.3172257 | -0.0647939 | 0.1798553 | -0.0647439 |
| 102654955 | ATP synthase subunit b, mitochondrial-like | 116.51647 | -0.4782838 | -0.0896692 | 0.3057816 | 0.4098934 |
| GB52790 | gamma-aminobutyric acid receptor-associated protein | 116.47545 | 0.1262545 | -0.0362937 | -0.1638312 | -0.0066064 |
| GB53421 | Golgi resident protein GCP60-like | 116.46662 | -0.0056854 | 0.0086737 | 0.1447571 | 0.0074084 |
| GB47409 | transmembrane protein 145-like isoform X1 | 116.36338 | -0.4348667 | -0.2311721 | 0.4665564 | 0.0690933 |
| GB41822 | acid phosphatase-like protein 2-like | 116.27882 | -0.4131980 | 0.2543138 | -0.0384230 | 0.0274197 |
| GB54702 | S1 RNA-binding domain-containing protein 1-like isoform X1 | 116.24635 | -0.3909295 | 0.1058792 | 0.1470224 | 0.0728667 |
| GB47306 | sulfhydryl oxidase 1-like | 116.12737 | -0.2877590 | -0.0311051 | 0.3053809 | 0.0075416 |
| 102654029 | uncharacterized protein LOC102654029 isoform X1 | 116.02257 | -0.4372411 | 0.0574868 | 0.1124620 | 0.9607665 |
| 726043 | leucine-rich repeat-containing protein 57-like | 115.98613 | -0.2208731 | 0.0799332 | 0.0560385 | -0.0141118 |
| GB50030 | SET and MYND domain-containing protein 5 | 115.90252 | 0.0860222 | -0.1231709 | -0.0246038 | -0.0270693 |
| GB40765 | ankyrin repeat domain-containing protein 12-like isoform X3 | 115.86309 | -0.6912359 | -0.1058212 | 0.0258948 | -0.0466013 |
| GB46752 | microtubule-associated serine/threonine-protein kinase-like, transcript variant X3 | 115.61251 | 0.3661762 | 0.2288786 | 0.0019782 | 0.1120890 |
| GB49621 | alpha-L-fucosidase | 115.48812 | -0.1944113 | -0.0561931 | -0.0319472 | 0.1204625 |
| GB41677 | adenosine kinase 1-like isoformX1 | 115.44721 | -0.0592447 | 0.0065554 | -0.0194049 | -0.0231758 |
| GB41510 | probable deoxyhypusine synthase-like isoform X1 | 115.40056 | -0.0703368 | -0.0234259 | 0.0914116 | 0.0207224 |
| GB44969 | lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial-like | 115.37107 | -0.0250647 | -0.0634631 | -0.0709718 | -0.2206717 |
| GB52803 | 28 kDa heat- and acid-stable phosphoprotein-like | 115.37050 | 0.3274790 | 0.0195979 | 0.1072625 | 0.0931148 |
| GB48059 | ecdysone receptor isoform B1 | 115.21224 | -0.4214454 | 0.1548010 | -0.0528363 | 0.1111929 |
| GB40654 | nuclear factor NF-kappa-B p110 subunit isoform X3 | 115.18235 | -0.4383493 | -0.0627767 | 0.3827163 | 0.0480067 |
| GB40032 | rRNA-processing protein UTP23 homolog | 115.16676 | -1.0839536 | 0.2009496 | 0.0477468 | 0.2661873 |
| GB47502 | uncharacterized protein LOC409690 isoform X2 | 115.13113 | -0.2990619 | -0.0561152 | 0.1506158 | -0.0359369 |
| GB44598 | threonine aspartase 1-like isoform X2 | 115.08758 | -0.1337951 | 0.0996834 | -0.0347439 | 0.2601827 |
| GB40431 | beta-ureidopropionase-like isoform 1 | 114.92861 | 0.4338961 | 0.0472415 | -0.0798943 | -0.0952375 |
| GB42693 | la protein homolog | 114.84560 | 0.0741635 | 0.1481442 | -0.1067247 | 0.0597770 |
| GB41545 | MD-2-related lipid-recognition protein-like | 114.83046 | 0.1911075 | -0.0147415 | 0.1202560 | -0.0734920 |
| GB48876 | probable methyltransferase-like protein 15 homolog isoform X1 | 114.68117 | -0.6777539 | -0.0687570 | 0.2094694 | 0.1353600 |
| GB49908 | cGMP-dependent protein kinase foraging | 114.55417 | 0.1960175 | 0.0596341 | 0.0978862 | -0.0087055 |
| GB51526 | uncharacterized protein LOC410093 isoform X1 | 114.44815 | -0.1162656 | 0.2363919 | -0.3980980 | 0.0133688 |
| GB55156 | THUMP domain-containing protein 1 homolog | 114.35872 | -0.5870861 | 0.0643893 | 0.0534351 | 0.1202009 |
| GB51963 | mitochondrial ribonuclease P protein 1 homolog | 114.33454 | -0.4909597 | -0.2002546 | -0.0138016 | 0.1394110 |
| GB40859 | uncharacterized protein LOC100576321 | 114.23306 | -0.6690510 | 0.0943930 | -0.6749086 | 0.1173962 |
| GB45133 | zinc finger protein 341-like isoform X2 | 114.07938 | 0.0964241 | 0.3366654 | 0.1240437 | -0.1139229 |
| GB53683 | integrator complex subunit 10-like isoform X3 | 114.07407 | 0.1911285 | -0.1438958 | 0.1709601 | 0.0591472 |
| 102655284 | serine–tRNA ligase, mitochondrial-like | 114.03910 | 0.1166727 | 0.0248453 | -0.0565676 | -0.0908400 |
| GB41117 | LOW QUALITY PROTEIN: serine/threonine-protein kinase 11-interacting protein-like | 114.01117 | -0.3404501 | -0.0612309 | 0.1694473 | -0.1730021 |
| GB52746 | zinc finger protein 845-like isoform X1 | 113.97888 | 0.2113675 | -0.0755249 | 0.1529030 | 0.1828755 |
| GB46790 | solute carrier family 35 member F5-like isoform X2 | 113.96927 | -0.0971515 | 0.0133984 | 0.1145284 | 0.1594460 |
| GB47820 | zinc finger protein 91-like isoform X2 | 113.94482 | -0.4734156 | 0.0495809 | 0.0593248 | 0.0541560 |
| GB54568 | uncharacterized protein LOC726916 | 113.94400 | -0.1930899 | 0.2523648 | 1.7874940 | 0.0423096 |
| GB48846 | organic cation transporter protein-like | 113.87596 | -0.8209230 | 0.1493236 | 0.0115209 | -0.2029515 |
| GB45150 | adenylate cyclase 3 isoform X2 | 113.84019 | -0.1187307 | 0.1818096 | -0.0377153 | -0.2268787 |
| GB54314 | nucleostemin 1 | 113.82965 | -0.5476981 | 0.1385254 | -0.2240878 | 0.0655058 |
| GB53824 | neuferricin-like | 113.76675 | -0.4587086 | -0.0437010 | 1.4207625 | -0.0185231 |
| GB41449 | cytoplasmic FMR1-interacting protein isoform 1 | 113.67719 | -0.0992664 | -0.0015260 | 0.0517477 | -0.0174742 |
| GB49497 | V-type proton ATPase subunit B-like | 113.56104 | 0.1135658 | -0.0710405 | 0.1697669 | -0.0183821 |
| GB48018 | uncharacterized protein LOC100578752 | 113.51308 | 0.1617925 | 0.0749814 | 0.0831486 | 0.1310039 |
| GB47418 | uncharacterized LOC409282, transcript variant X3 | 113.42803 | -0.1838162 | 0.0000321 | 0.1802775 | 0.0874728 |
| GB45318 | uncharacterized protein LOC725813 | 113.41338 | 0.0274879 | -0.1252128 | -0.0095494 | 0.0988733 |
| GB42083 | uncharacterized protein LOC100578864 | 113.32399 | -1.0180120 | -0.0754823 | 0.7138082 | -0.0769110 |
| GB45041 | metallophosphoesterase 1 homolog | 113.21960 | -0.2034999 | 0.0398818 | 2.1048537 | 0.1223707 |
| GB41378 | probable helicase with zinc finger domain-like | 113.17932 | -0.3077168 | 0.2912704 | 0.0237014 | -0.0556308 |
| GB41259 | INO80 complex subunit B-like isoform X1 | 113.15565 | -0.3103358 | 0.0697064 | 0.0526251 | 0.1007072 |
| GB49235 | WD repeat-containing protein 24-like | 113.03888 | -0.0494100 | 0.0898371 | 0.1022128 | 0.0346863 |
| GB46590 | interaptin-like | 112.99967 | -0.2683662 | 0.0895455 | 0.0467887 | 0.1232528 |
| GB52697 | origin recognition complex subunit 4 isoform X2 | 112.94878 | -0.7997903 | -0.1736332 | 0.0284677 | -0.0538377 |
| GB46267 | uncharacterized protein LOC727260 | 112.86594 | 0.3741879 | 0.1307184 | -0.0529877 | 0.1986751 |
| GB54729 | ATP-dependent DNA helicase PIF1-like | 112.83696 | -0.8082459 | 0.2369653 | -0.0595648 | -0.4012492 |
| GB54256 | uncharacterized protein LOC100577343 isoform X2 | 112.82018 | -0.2315563 | -0.2971905 | 0.2461804 | 0.0149778 |
| GB40075 | importin subunit beta-1 isoform X2 | 112.81441 | 0.0097743 | 0.2466988 | -0.2500319 | -0.1020705 |
| GB41671 | uncharacterized protein LOC724294 | 112.79739 | -0.4648615 | -0.0299269 | -0.0278379 | 0.0200333 |
| GB55187 | conserved oligomeric Golgi complex subunit 6-like | 112.73256 | 0.0051348 | 0.0544917 | 0.0988191 | 0.0578531 |
| GB49427 | protein RRNAD1-like | 112.69993 | -0.2562656 | 0.0326873 | -2.0529899 | 0.1670561 |
| GB40296 | kanadaptin-like | 112.64401 | -0.5470967 | 0.0365528 | 0.0502521 | 0.0936168 |
| GB42785 | spermatogenesis-defective protein 39 homolog | 112.60368 | -0.1285075 | 0.0191215 | -0.1504102 | 0.1890441 |
| GB41291 | heparan-alpha-glucosaminide N-acetyltransferase-like isoform X2 | 112.56289 | 0.0145168 | -0.1333306 | 0.1518642 | 0.0935770 |
| GB53296 | pipsqueak | 112.53499 | -0.1228549 | 0.1778994 | 0.0948562 | 0.0356113 |
| GB55601 | protein YIPF1-like | 112.49040 | 0.1034192 | -0.0577733 | 0.1365281 | 0.1102380 |
| GB46714 | nucleoporin SEH1 isoform X1 | 112.47935 | 0.4605465 | 0.0223960 | 0.1280122 | 0.0693529 |
| GB45179 | mRNA-capping enzyme | 112.42404 | 0.2079675 | -0.0086639 | 0.0046254 | -0.0884306 |
| GB42697 | SPRY domain-containing protein 7-like isoform 1 | 112.33314 | 0.3623415 | -0.1559320 | 0.1265800 | 0.2416789 |
| GB55112 | histone H4 transcription factor-like isoform X1 | 111.95090 | -0.5402079 | 0.0297658 | -0.3128531 | -0.1327502 |
| GB52804 | lysophospholipid acyltransferase 7-like | 111.89353 | -0.3367843 | 0.0934704 | 0.6904438 | -0.0688806 |
| GB50024 | origin recognition complex subunit 1 | 111.89055 | -0.1826989 | 0.3541047 | 0.1395557 | 0.1876799 |
| 100578218 | mimitin, mitochondrial-like | 111.86932 | -0.2705291 | 0.0416336 | 0.0851672 | 0.0903698 |
| GB45372 | kynurenine formamidase-like isoform X4 | 111.85022 | -0.1700784 | -0.0106097 | 0.0115557 | -0.0291510 |
| GB42814 | calcium-transporting ATPase type 2C member 1-like isoform X2 | 111.66361 | 0.1504158 | 0.0404337 | -0.2866773 | -0.0700889 |
| 727332 | protein BUD31 homolog isoform X1 | 111.66102 | 0.4063355 | -0.0614017 | 0.1207100 | 0.0113972 |
| GB52455 | protein CASC4-like isoform X2 | 111.64079 | 0.3473047 | -0.0645021 | -0.0416983 | -0.1037842 |
| GB45740 | 39S ribosomal protein L40, mitochondrial | 111.62270 | -0.1452949 | 0.0202724 | -0.1746363 | 0.0713631 |
| GB54326 | apoptosis regulatory protein Siva-like | 111.41669 | -0.4168160 | 0.0987478 | 0.0397571 | -0.0625042 |
| GB52701 | chloride channel protein 2-like isoform X3 | 111.30866 | -0.2212543 | -0.1462758 | 0.2926829 | -1.4150749 |
| GB42664 | probable ATP-dependent RNA helicase DDX43-like | 111.28840 | -0.2010126 | 0.1264142 | -0.1278858 | 0.1035423 |
| GB55010 | spermine synthase-like isoformX1 | 111.22158 | 0.1096710 | 0.0019226 | -0.7507208 | 0.0338901 |
| 410849 | protein sly1 homolog | 111.16053 | -0.1188614 | 0.0375003 | -0.1037166 | -0.2413957 |
| GB42820 | N-alpha-acetyltransferase 30-like | 111.12459 | -0.4079194 | 0.0770459 | 0.0675232 | 0.1604197 |
| GB45185 | FAS-associated factor 2 | 110.99829 | 0.1199888 | -0.0725345 | 0.3138127 | -0.1180144 |
| GB50146 | xaa-Pro aminopeptidase 1-like isoform X1 | 110.99316 | -0.0492869 | 0.1650069 | 0.0234085 | -0.0524205 |
| GB53794 | melanotransferrin | 110.85805 | 0.3004488 | -0.0245872 | 0.0582367 | -0.1024655 |
| GB45749 | sorting nexin-4-like isoform X1 | 110.82238 | 0.2778389 | -0.1776120 | 0.0937926 | 0.0809841 |
| GB43376 | low molecular weight phosphotyrosine protein phosphatase-like isoform X2 | 110.78268 | -0.1331990 | 0.0282981 | -0.0199650 | 0.2658863 |
| GB52982 | magnesium transporter NIPA2 | 110.71762 | -0.1153847 | -0.0480350 | 0.1549438 | 0.1172079 |
| GB47735 | endonuclease III-like protein 1-like | 110.66376 | -0.1849047 | 0.1516198 | 2.4414161 | 0.2298213 |
| GB53846 | structural maintenance of chromosomes protein 5-like | 110.62516 | -0.1045349 | -0.1549860 | -0.0174664 | 0.0092808 |
| GB47879 | phosphatidylinositol 4-kinase alpha-like | 110.58308 | 0.1399439 | -0.0525335 | 0.5625816 | 0.0484953 |
| GB40030 | adenylyltransferase and sulfurtransferase MOCS3-like | 110.32384 | -0.3108809 | -0.0444118 | 0.0700305 | 0.1701809 |
| GB44745 | zinc finger protein 267-like | 110.27017 | -0.5940097 | 0.1307057 | -0.0474234 | -0.1270068 |
| GB49894 | probable cytochrome P450 6a14 isoform X1 | 110.20932 | -0.4231279 | 0.2240414 | 0.3679933 | 0.1745154 |
| GB49171 | protein sidekick isoformX2 | 110.18850 | -0.1128901 | 0.1588067 | -0.7407300 | 0.3438187 |
| GB42707 | mortality factor 4-like protein 1-like isoform X1 | 110.15387 | -0.1135430 | 0.0153714 | -0.0186611 | 0.1485030 |
| GB53681 | cytochrome b5 reductase 4-like isoform X4 | 110.08065 | -0.2111228 | -0.1133910 | 0.0298355 | -0.0186916 |
| GB40673 | lambda crystallin-like protein | 110.04641 | 0.0347234 | -0.0267619 | -0.2175415 | 0.0314696 |
| GB53376 | uncharacterized protein C18orf8-like | 109.92071 | 0.1849792 | 0.0815656 | 0.0507849 | 0.2440196 |
| GB41161 | probable DNA-directed RNA polymerases I and III subunit RPAC2-like | 109.87981 | -0.0528962 | -0.0188192 | 0.0057755 | -0.0011473 |
| GB41335 | N(6)-adenine-specific DNA methyltransferase 2-like | 109.87882 | 0.0246216 | -0.0172813 | 0.1078644 | 0.1227047 |
| 410675 | protein BTG3-like isoform X2 | 109.72165 | 0.2136615 | -0.1669195 | 0.0877332 | 0.0036217 |
| GB46477 | LOW QUALITY PROTEIN: putative inositol monophosphatase 3-like | 109.69954 | 0.0835631 | -0.1117051 | 0.0823936 | 0.1410098 |
| GB48017 | transforming growth factor beta regulator 1-like | 109.68454 | -0.0015045 | 0.0281990 | -0.0783565 | 0.2286131 |
| GB51160 | ralBP1-associated Eps domain-containing protein 2-like isoform X1 | 109.63895 | 0.0070839 | -0.1034523 | -0.0198323 | 0.0928778 |
| 102656137 | coiled-coil domain-containing protein 174-like | 109.58872 | -0.5943258 | 0.0142894 | 0.1179950 | 0.1556611 |
| GB41654 | deformed epidermal autoregulatory factor 1 | 109.49718 | 0.2245943 | 0.1382421 | 0.0524434 | -0.0791638 |
| GB54659 | transcription initiation factor TFIID subunit 8-like | 109.44000 | -0.0687825 | 0.1108418 | -0.0510725 | 0.1920184 |
| GB50993 | DNA-directed RNA polymerase III subunit RPC4-like | 109.43993 | 0.1217160 | 0.1176160 | 0.0616132 | 0.2109731 |
| GB55164 | lethal(2) giant larvae protein homolog 1-like isoform X9 | 109.41728 | 0.1167371 | -0.0136240 | 0.0593902 | 0.5008095 |
| GB55463 | caprin homolog | 109.28031 | 0.1306977 | 0.1549545 | 0.0043409 | -0.3999232 |
| GB52975 | uncharacterized protein LOC409658 | 109.21241 | -0.0273347 | 0.1067078 | 0.0021505 | -0.1274968 |
| GB47162 | ADP-ribosylation factor-like protein 13B-like isoform X2 | 109.13613 | -0.2809829 | -0.0087423 | 0.0874894 | -0.0487411 |
| GB51675 | uncharacterized protein R102.4-like isoform X2 | 108.94504 | -0.2821518 | 0.0539370 | -0.0423334 | 0.0714882 |
| GB41982 | polycomb protein Scm isoform X2 | 108.78823 | 0.2169077 | -0.0011055 | 0.0656385 | -0.1727408 |
| GB48031 | vacuolar protein sorting-associated protein 54 | 108.69785 | -0.2016125 | -0.1654271 | 0.0476317 | -0.0396671 |
| GB40400 | tyrosine-protein kinase PR2-like | 108.54632 | -0.2171005 | -0.4280187 | -0.4622659 | 0.5577777 |
| GB47295 | KH domain-containing protein C56G2.1-like isoform X3 | 108.46357 | -0.6147302 | 0.1431407 | -0.0754483 | -0.0953102 |
| GB44558 | probable malonyl-CoA-acyl carrier protein transacylase, mitochondrial-like | 108.44477 | -0.4227646 | -0.0823538 | -0.0371695 | -0.0620607 |
| GB47310 | NECAP-like protein CG9132-like isoform X1 | 108.36423 | -0.3260422 | -0.0189028 | -0.0415143 | -0.2137695 |
| GB40489 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | 108.33979 | -0.5737909 | -0.0697277 | 0.1215298 | 0.1709151 |
| GB53256 | 39S ribosomal protein L3, mitochondrial | 108.33198 | -0.0168834 | -0.0345604 | -0.0077669 | 0.2118512 |
| GB51743 | protein decapentaplegic | 108.31694 | -0.1392645 | 0.0101274 | 0.2936252 | -0.0528951 |
| 102656258 | nuclear RNA export factor 2-like | 108.13832 | 0.2240183 | -0.0589057 | -0.0819122 | 0.0338447 |
| GB42779 | alpha-1,3/1,6-mannosyltransferase ALG2-like | 108.13632 | -0.3617389 | 0.0649762 | -0.1834664 | 0.0944667 |
| GB45515 | uncharacterized protein LOC411721 | 108.09928 | -0.4927600 | 0.0402670 | 0.1585531 | 0.1398947 |
| GB43270 | MATH and LRR domain-containing protein PFE0570w-like isoform X2 | 108.05702 | -0.8636913 | -0.0222576 | 0.1897120 | -0.4373773 |
| GB44877 | probable 39S ribosomal protein L49, mitochondrial-like | 108.02184 | 0.1149562 | 0.0881126 | 0.0710677 | 0.0947513 |
| GB53606 | histone-lysine N-methyltransferase eggless | 107.88955 | 0.2852783 | -0.0166182 | 0.2152273 | -0.0655172 |
| GB47885 | probable cytochrome P450 304a1 | 107.80353 | 0.0398263 | -0.1050613 | -0.2240712 | 0.4697969 |
| 102656433 | DNA ligase 1-like | 107.74829 | -0.3700309 | 0.1291451 | 0.0716274 | 0.0333112 |
| GB53021 | PR domain zinc finger protein 10-like isoform X2 | 107.72912 | -0.2280685 | 0.0258377 | 0.2229598 | -0.0857481 |
| GB49067 | zinc finger protein GLI1-like | 107.63472 | -0.1125468 | -0.0854113 | 0.0867423 | -0.0385636 |
| GB48158 | uncharacterized protein LOC408905 | 107.59322 | 0.0455199 | 0.2143792 | -0.5167773 | 0.1506558 |
| GB43353 | uncharacterized protein LOC727578 | 107.58625 | -0.2261745 | -0.0599955 | 0.3144350 | 0.0753310 |
| GB43586 | uncharacterized protein LOC100576934 | 107.50198 | -0.0264041 | -0.1218398 | 0.1726393 | -0.0034741 |
| GB49139 | autophagy protein 5 | 107.49715 | 0.0630807 | 0.0000015 | 1.2160955 | 0.0953454 |
| GB54253 | uncharacterized protein LOC727266 | 107.46021 | 0.0966828 | -0.0487246 | 0.1369556 | -0.0515740 |
| GB55071 | F-box only protein 9-like | 107.38499 | -0.1974416 | -0.1360946 | 0.1840697 | 0.0962722 |
| GB42139 | E3 ubiquitin-protein ligase RFWD3 | 107.11235 | -0.1873586 | 0.0553139 | -0.0806206 | 0.1609753 |
| GB52058 | chromobox protein homolog 1-like isoform X1 | 107.04485 | 0.0787815 | 0.1282460 | 0.2087798 | 0.1244195 |
| GB52888 | uncharacterized protein C9orf85 homolog | 106.98007 | -0.4439181 | -0.0639822 | 0.0782403 | 0.1518754 |
| 102655203 | EKC/KEOPS complex subunit TPRKB-like | 106.91963 | -0.4573536 | -0.2727266 | 1.2798976 | 0.1655323 |
| GB40949 | homologous-pairing protein 2 homolog | 106.91839 | -0.1636276 | -0.0494989 | -0.0323305 | 0.2030138 |
| GB47461 | protein inturned-like | 106.87189 | -0.4167487 | 0.0531368 | -0.0379586 | 0.2089070 |
| GB50796 | probable N-acetyltransferase CML3-like | 106.86955 | -0.0095568 | -0.0936732 | 0.1011395 | -0.0896497 |
| GB49050 | zinc finger protein 484-like isoform X2 | 106.82084 | -0.2077000 | 0.0962310 | -0.1464946 | 0.0013437 |
| GB41176 | polyglutamine-binding protein 1-like | 106.73765 | -0.3465634 | 0.1348326 | 1.1815555 | 0.0107641 |
| GB41889 | aminoacylase-1-like | 106.69130 | 0.1628694 | 0.0439385 | -0.0517544 | -0.2290878 |
| GB52266 | furin-like protease 2-like | 106.59146 | -0.9273815 | -0.0739821 | 0.0110234 | 0.0435829 |
| GB52642 | probable serine/threonine-protein kinase tsuA-like | 106.36980 | -0.0448327 | -0.1595186 | -0.0659380 | -0.0027964 |
| GB54808 | protein maelstrom homolog isoform X3 | 106.35030 | -0.1166352 | 0.1758062 | 0.0510711 | -0.0130529 |
| GB48255 | isochorismatase domain-containing protein 2, mitochondrial-like isoform X3 | 106.24834 | 0.0918452 | 0.0040123 | -0.1468156 | 0.0018809 |
| GB46792 | inosine-5’-monophosphate dehydrogenase isoform 1 | 106.19775 | 0.4914342 | -0.0111189 | 0.0485542 | -0.1913066 |
| GB55076 | signal recognition particle 19 kDa protein | 106.10218 | 0.2028129 | -0.0745961 | 0.0349813 | -0.0756541 |
| GB48414 | trafficking protein particle complex subunit 3 | 106.01394 | -0.0169477 | 0.1372539 | 0.0776237 | 0.0590079 |
| GB54387 | probable ribosome biogenesis protein RLP24-like | 106.01234 | 0.2373581 | 0.1044930 | -0.0700100 | 0.1311054 |
| GB50996 | DNA-directed RNA polymerase II subunit RPB3-like | 105.92469 | 0.0411047 | 0.2190810 | 0.1506329 | 0.0085849 |
| GB46476 | ubiquitin-conjugating enzyme E2 W-like | 105.84266 | 0.1427913 | 0.0311380 | 0.0333240 | 0.0490065 |
| GB50298 | putative transferase CAF17 homolog, mitochondrial-like | 105.80257 | -0.1317539 | 0.0136390 | -0.0400881 | -0.0536425 |
| GB47389 | LETM1 and EF-hand domain-containing protein anon-60Da, mitochondrial-like | 105.79887 | -0.0238946 | 0.0791973 | -0.1495645 | 0.1909181 |
| GB50103 | crossover junction endonuclease EME1-like | 105.72601 | -0.2979254 | -0.0526824 | 0.2681103 | -0.0163311 |
| GB44705 | mediator of RNA polymerase II transcription subunit 6 | 105.62122 | 0.0051881 | 0.0372643 | 0.2047516 | 0.0110930 |
| GB40843 | leucine-rich repeat-containing protein 47-like | 105.45903 | -0.0414463 | -0.0918743 | 0.0518263 | 0.0550407 |
| GB42384 | serine/threonine-protein kinase RIO2-like | 105.24271 | -0.2352432 | 0.1476477 | 0.3265530 | 0.0310779 |
| GB41171 | uncharacterized protein LOC551860 | 105.19001 | -0.1053643 | 0.3128545 | -0.1476829 | 0.3698557 |
| GB51627 | dehydrodolichyl diphosphate synthase-like isoform X4 | 105.12078 | -0.9931325 | 0.0398059 | -0.0781198 | 0.2136125 |
| GB43495 | tRNA dimethylallyltransferase, mitochondrial-like isoform X2 | 105.11658 | -0.2413611 | 0.1205191 | 0.0740874 | -0.0059074 |
| GB40355 | coiled-coil domain-containing protein 94-like isoform X2 | 105.11203 | -0.5710464 | 0.2660566 | -0.0090931 | -0.0175476 |
| 413881 | AP-3 complex subunit sigma-2-like, transcript variant X2 | 105.07424 | 0.1559031 | 0.0272858 | 0.1817562 | 0.0218726 |
| GB46697 | carbonic anhydrase 2 | 105.02529 | -0.2196129 | -0.0295819 | -0.1057319 | -0.0084071 |
| GB47961 | mitogen-activated protein kinase kinase kinase 4 | 105.01370 | 0.1228679 | -0.0480623 | 0.1114288 | -0.3113776 |
| GB47454 | putative inhibitor of apoptosis | 104.96798 | -0.4327730 | -0.0272042 | -0.3857389 | 0.0929237 |
| GB45389 | E3 ubiquitin-protein ligase RNF123-like isoform X1 | 104.88151 | -0.4752533 | -0.1062509 | 0.0259576 | -0.2148111 |
| GB49451 | mediator of RNA polymerase II transcription subunit 28-like | 104.79311 | 0.2205252 | -0.0148582 | 0.0862162 | 0.0643483 |
| GB55518 | uncharacterized protein C17orf59 homolog isoform X3 | 104.79100 | -0.0004063 | 0.0054668 | 0.2120552 | 0.1971523 |
| GB40515 | myotubularin-related protein 4-like isoform X1 | 104.53951 | 0.1288438 | 0.0562030 | 0.1026152 | -0.0126216 |
| 551412 | polycomb protein EED-like | 104.50265 | 0.1029125 | 0.2716119 | -0.0025489 | 0.0039970 |
| GB40862 | UPF0545 protein C22orf39 homolog | 104.44706 | -0.2674479 | -0.1062744 | 0.0962665 | 0.2284805 |
| GB44358 | leucine-rich repeat-containing protein 48-like isoform X1 | 104.42516 | -1.1548688 | -0.5974715 | 0.0206248 | 0.3427824 |
| GB45083 | protein FRG1 homolog | 104.39137 | -0.2219969 | 0.0667667 | 0.1259454 | 0.1407993 |
| GB47834 | histone deacetylase complex subunit SAP30 homolog isoform X1 | 104.35493 | -0.1854688 | -0.0155802 | -0.9767427 | -0.1757420 |
| GB45193 | glutaryl-CoA dehydrogenase, mitochondrial | 104.32422 | 0.1194852 | -0.0100388 | 0.0610759 | -0.1818264 |
| GB49553 | stromal membrane-associated protein 1-like | 104.27586 | 0.0843935 | 0.0394309 | 0.2842563 | 0.0822133 |
| GB45020 | uncharacterized protein LOC408364 isoform X1 | 104.15960 | -0.0178876 | -0.0032009 | 0.1743422 | 0.1138339 |
| GB43627 | zinc finger protein GLI2-like isoform X4 | 103.99231 | -0.5250875 | 0.0502684 | 0.1829646 | 0.0556758 |
| GB45025 | mTERF domain-containing protein 1, mitochondrial-like | 103.95754 | -0.7229887 | -0.1257126 | 0.4072139 | 0.2728811 |
| GB42089 | transmembrane protein 256 homolog | 103.93478 | 0.0788001 | -0.0391551 | -0.1115476 | 0.0284092 |
| GB41569 | UPF0364 protein C6orf211 homolog isoform X1 | 103.88984 | -0.3512359 | -0.2315017 | 0.0022055 | 0.1545650 |
| GB47443 | mesoderm induction early response protein 1-like isoform X6 | 103.82427 | -0.1561379 | 0.0974842 | 0.2702222 | -0.0396145 |
| GB50809 | sprT-like domain-containing protein Spartan-like isoform X3 | 103.72201 | -0.0952836 | 0.0350733 | -0.0007735 | -0.0329958 |
| GB42805 | heparan-sulfate 6-O-sulfotransferase 2 | 103.64416 | 0.0473980 | -0.0911816 | 0.0202213 | 0.0471005 |
| GB50167 | probable 28S ribosomal protein S26, mitochondrial | 103.63404 | -0.8383698 | 0.5278059 | 0.0847339 | 0.1749817 |
| 725620 | 28S ribosomal protein S33, mitochondrial | 103.50080 | -0.2003903 | -0.0319375 | 0.0753086 | 0.1375421 |
| GB53672 | failed axon connections isoform X2 | 103.41105 | 0.2266880 | 0.1568684 | 0.0240717 | -2.4708213 |
| GB50694 | F-box/LRR-repeat protein 20-like isoform X2 | 103.39893 | -0.1127084 | -0.0916047 | 0.1505171 | 0.0054693 |
| GB43126 | transmembrane protein 184B-like isoform X1 | 103.38120 | -0.0081802 | 0.0111398 | 0.1702457 | -0.0576392 |
| GB48126 | putative uncharacterized protein DDB_G0282133-like | 103.29270 | -0.1005232 | 0.1466956 | 0.0270128 | -0.2610919 |
| GB51465 | NAD-dependent protein deacetylase Sirt7 | 103.14614 | -0.2175926 | 0.3000467 | -0.0779014 | 0.0510026 |
| GB53430 | reticulon-4-interacting protein 1, mitochondrial-like isoform 1 | 103.12063 | 0.0618704 | 0.0091431 | 0.0516170 | 0.2320996 |
| GB49837 | WD repeat domain phosphoinositide-interacting protein 3-like isoform 1 | 103.08680 | 0.3960110 | 0.0759265 | 0.0977843 | 0.0262251 |
| GB41157 | RPII140-upstream gene protein-like | 102.92500 | -0.4811127 | -0.0234584 | 1.2732615 | 0.1490042 |
| 102654398 | DNA repair protein xrcc4-like | 102.91716 | -1.0823790 | 0.0229883 | 0.3274392 | 0.2404575 |
| GB55902 | cystathionine-beta-synthase | 102.90506 | -0.0182529 | 0.0636398 | -0.0983190 | -0.0632421 |
| GB41030 | UV excision repair protein RAD23 homolog B-like isoformX2 | 102.87269 | 0.1722687 | -0.0654878 | -0.1117246 | 0.0523045 |
| GB52091 | protein suppressor of forked-like isoform X3 | 102.86446 | 0.1621853 | -0.0201628 | 0.1646234 | -0.0608151 |
| 102655971 | 5’-nucleotidase domain-containing protein 1-like | 102.76983 | 0.1451592 | -0.0678287 | 0.0949727 | 0.1648122 |
| GB44695 | lysophospholipase-like protein 1-like isoform X1 | 102.71681 | -0.0722567 | -0.0606883 | -0.0222535 | 0.1276303 |
| GB43434 | enolase-phosphatase E1-like isoform 2 | 102.71007 | -0.0490663 | 0.1371099 | 0.0659162 | 0.2804967 |
| 724133 | peptide chain release factor 1-like, mitochondrial-like | 102.70530 | -0.2378289 | 0.1720038 | 0.1298950 | 0.0423699 |
| 102656263 | protein farnesyltransferase subunit beta-like | 102.68251 | 0.0024406 | -0.0462655 | 0.0333747 | -0.1234934 |
| GB49242 | apoptosis-inducing factor 3 isoform X1 | 102.66647 | 0.1783949 | -0.1737989 | 0.1272648 | 0.0426385 |
| GB53201 | 39S ribosomal protein L44, mitochondrial | 102.66201 | -0.4322023 | 0.0010946 | -0.0754102 | 0.2410727 |
| GB41441 | ubiquitin-like protein 3-like isoform X3 | 102.65399 | -0.3155675 | 0.0910099 | -0.1761630 | 0.2201493 |
| GB51756 | zinc finger protein 69-like isoform X1 | 102.56874 | 0.3428832 | -0.1067140 | 0.0444482 | -0.1396647 |
| GB40146 | GTP-binding protein ypt7 | 102.54877 | 0.3700556 | -0.1190567 | 0.1202566 | 0.0782783 |
| GB55283 | uncharacterized protein LOC552036 isoform X2 | 102.50757 | 0.1742628 | -0.0124449 | 0.1809800 | 0.1175203 |
| GB44540 | DNA repair protein RAD51 homolog 1 | 102.49657 | 0.1046023 | 0.2095263 | -0.2533528 | 0.0265824 |
| GB51281 | uncharacterized protein LOC409514 isoform 1 | 102.48684 | -0.1879763 | -0.2360590 | 0.1539353 | -0.9075939 |
| GB51080 | uncharacterized protein C16orf52 homolog A isoform X4 | 102.46539 | 0.4100117 | 0.0719983 | 0.1228402 | 0.0834943 |
| 408979 | eukaryotic translation initiation factor 5 isoform X2 | 102.32904 | 0.1148310 | 0.1468431 | -0.0138056 | 0.1650226 |
| GB52473 | DNA polymerase eta | 102.11287 | 0.0153179 | 0.1884311 | 0.0741648 | 0.1919675 |
| GB53916 | eukaryotic translation initiation factor 3 subunit E-like isoform X2 | 102.08028 | 0.0699058 | 0.0878676 | -0.0374221 | -0.5740508 |
| GB46495 | tetratricopeptide repeat protein 4 | 101.98783 | -0.2649464 | 0.0456088 | -0.0278906 | 0.1411271 |
| GB54981 | cysteine-rich hydrophobic domain 2 protein-like | 101.93489 | -0.0748597 | 0.0227370 | 0.0241328 | -0.0051259 |
| GB40648 | disks large 1 tumor suppressor protein-like, transcript variant X2 | 101.91669 | -0.3241854 | -0.1897401 | 0.3085009 | 0.1043669 |
| GB55452 | apolipophorin-III-like protein precursor | 101.76044 | -0.1917584 | -0.0045336 | -0.1298917 | -0.1635547 |
| GB42941 | uridine-cytidine kinase-like 1-like isoform 1 | 101.72984 | 0.0976224 | -0.0348647 | 0.1663574 | 0.0641137 |
| 102654889 | zinc finger protein 235-like | 101.70459 | -0.7249597 | 0.1640394 | 0.3733752 | -0.1871758 |
| GB45281 | E3 ubiquitin-protein ligase hyd isoform X2 | 101.69504 | 0.0323415 | 0.0878511 | 0.0713970 | -0.1985058 |
| GB46986 | 39S ribosomal protein L46, mitochondrial | 101.68460 | -0.2538520 | 0.0349140 | -0.0514653 | 0.3188129 |
| GB45665 | fidgetin-like protein 1-like | 101.62753 | -0.0678293 | 0.2886192 | -0.0245314 | -0.0428502 |
| GB46067 | putative N-acetylglucosamine-6-phosphate deacetylase | 101.48716 | 0.3961385 | -0.0088652 | 0.1103986 | 0.2691109 |
| GB47719 | protein KRI1 homolog | 101.43537 | -0.3082972 | 0.0751623 | -0.0139878 | 0.0254988 |
| GB43552 | leucine-rich repeat-containing protein 15-like isoform X1 | 101.27937 | -0.2382482 | -0.1407269 | -0.0256372 | 0.0864164 |
| GB45618 | E3 ubiquitin-protein ligase mind-bomb isoform X3 | 101.18146 | -0.4128355 | 0.0661485 | 0.0759655 | -0.0962491 |
| GB53790 | sorting nexin-27-like isoform X3 | 101.16913 | -0.5244638 | 0.0567308 | 0.0265912 | -0.0183979 |
| GB44734 | checkpoint protein HUS1-like isoform X1 | 101.10484 | -0.9468876 | 0.3714348 | -0.0354591 | 0.4284949 |
| GB49349 | CTL-like protein 1-like | 101.08734 | -0.0257116 | -0.2350726 | 0.0516107 | 0.0343246 |
| GB41153 | peroxisomal membrane protein PEX16-like | 100.92772 | -0.3174647 | 0.2076297 | -0.0068948 | 0.0130889 |
| GB50197 | splicing factor 3A subunit 3 isoform X1 | 100.81051 | 0.0513399 | 0.1173138 | 0.2056825 | 0.1779810 |
| GB49172 | nucleosome-remodeling factor subunit NURF301-like isoform X2 | 100.80501 | -0.4133487 | -0.0845701 | 0.0148700 | -0.1943096 |
| GB46681 | cyclin-K | 100.73121 | 0.1702849 | -0.0021017 | 0.0137749 | -0.0616683 |
| GB51000 | alpha-(1,6)-fucosyltransferase | 100.68020 | -0.0189872 | 0.0752420 | -0.0050330 | 0.1634025 |
| 100578779 | uncharacterized protein LOC100578779 | 100.66240 | -0.8562807 | -0.1249181 | 0.2185429 | 0.0369246 |
| GB44113 | 5’-AMP-activated protein kinase subunit beta-1 isoform X2 | 100.53836 | 0.1717533 | -0.0223231 | 0.0052793 | 0.0651376 |
| GB44611 | digestive organ expansion factor homolog | 100.48490 | 0.1137899 | 0.0334740 | 0.0235002 | -0.0499674 |
| GB51117 | rho GTPase-activating protein 18-like isoform X2 | 100.43732 | -0.4802457 | 0.0563335 | 0.0583868 | -0.0174060 |
| GB46132 | glycosaminoglycan xylosylkinase-like | 100.40167 | 0.6697098 | 0.0066428 | 0.2236263 | 0.0427335 |
| GB53415 | WW domain-binding protein 2-like isoform X1 | 100.37013 | 0.4324750 | -0.0179538 | -0.4569268 | -0.3257528 |
| GB51060 | probable serine/threonine-protein kinase DDB_G0282963 isoform X4 | 100.34867 | -1.7290084 | 0.0450148 | -0.2720275 | 0.5677084 |
| GB46877 | kinase suppressor of Ras 2 | 100.34579 | 0.1500412 | 0.1318707 | 0.1017462 | 0.1184538 |
| GB41215 | rotatin-like isoform X1 | 100.30869 | -0.1119417 | -0.1846935 | 0.7596468 | 0.1547873 |
| GB17746 | putative lipoyltransferase 2, mitochondrial | 100.23752 | -0.4370499 | -0.0097135 | -0.6321628 | 0.1179569 |
| 102656846 | G2/mitotic-specific cyclin-A-like | 100.17858 | 0.1031448 | 0.3257691 | -0.1341649 | 0.1746287 |
| GB44776 | lanC-like protein 2-like isoform X5 | 100.10728 | 0.0568897 | 0.0117974 | 0.1923044 | -0.0269398 |
| GB53029 | pre-mRNA-processing factor 17 isoform X1 | 100.09112 | -0.0270357 | 0.2165522 | -0.1192458 | -0.1265628 |
| GB49012 | glycerol kinase isoform X1 | 99.98457 | 0.2732696 | 0.2403865 | 0.0729685 | 0.0307508 |
| GB41794 | sorting nexin-30-like | 99.88747 | 0.0074558 | -0.0904362 | 0.0466807 | 0.1762950 |
| 102655841 | uncharacterized protein LOC102655841 | 99.84417 | 0.1915649 | -0.0426754 | 0.0551005 | 0.1113893 |
| GB48963 | chromosome transmission fidelity protein 18 homolog isoform X2 | 99.75002 | -1.3593902 | 0.0638973 | 1.1600603 | 0.2011274 |
| GB55864 | UDP-glucuronosyltransferase 1-8-like | 99.64440 | -0.5004614 | 0.0806789 | -0.0497709 | 0.0472375 |
| GB53710 | breakpoint cluster region protein-like isoform X3 | 99.60728 | -0.0073350 | -0.1310536 | 0.0053834 | -0.1152807 |
| GB42383 | uncharacterized protein MAL13P1.304-like | 99.58586 | -1.0495232 | 0.0604998 | -0.1394046 | 0.2128702 |
| GB45228 | chondroitin sulfate synthase 2-like | 99.56951 | -0.5713597 | 0.0558316 | 0.0990164 | 0.0258455 |
| GB49709 | coiled-coil domain-containing protein 86-like | 99.49929 | -0.0832927 | 0.1820437 | -0.0467071 | -0.0246030 |
| GB42650 | GPI mannosyltransferase 4-like | 99.40417 | -0.5355793 | 0.0578004 | 0.1013676 | 0.0982105 |
| GB42844 | guanine nucleotide exchange factor DBS-like isoform X3 | 99.39893 | -0.3899928 | -0.2577590 | 0.1891873 | -0.0036907 |
| GB47802 | signal recognition particle receptor subunit beta | 99.38453 | 0.2915627 | 0.0393262 | 0.1650618 | 0.1204635 |
| GB53403 | uncharacterized LOC100577196, transcript variant X4 | 99.30518 | 0.1477353 | 0.1130084 | -0.0058811 | 0.2445137 |
| GB48022 | protein henna-like isoform X3 | 99.27433 | -0.3617271 | -0.3258998 | -0.0630637 | 0.1086631 |
| GB54698 | DNA-directed RNA polymerase I subunit RPA49-like isoform X3 | 99.03684 | -0.5437046 | 0.0441890 | -0.1593698 | 0.0777006 |
| GB52456 | LOW QUALITY PROTEIN: protein DENND6A-like | 98.90104 | 0.2415306 | 0.2165214 | 0.1519922 | 0.0348819 |
| GB42356 | arginine-glutamic acid dipeptide repeats protein-like | 98.80861 | -0.6745294 | -0.0959205 | 0.1892585 | 0.0415860 |
| GB55020 | plexin-A4 isoform X3 | 98.80230 | -0.0434397 | -0.0903060 | 0.0739980 | -0.2630989 |
| GB48708 | sphingomyelin synthase-related 1 | 98.75182 | -0.0999252 | -0.2259788 | 0.0009290 | -0.0348003 |
| 727539 | lysocardiolipin acyltransferase 1-like | 98.71258 | 0.1633727 | 0.0217869 | -0.0510797 | -0.0376360 |
| GB54927 | BRCA1-associated protein-like isoform X2 | 98.69078 | -0.1718279 | -0.0135477 | 0.1312334 | 0.0790459 |
| GB41029 | cell cycle control protein 50A-like isoform X5 | 98.59469 | -0.1301546 | -0.0304659 | 0.2460747 | 0.0564584 |
| GB44149 | proclotting enzyme isoform X1 | 98.59213 | -0.1733514 | -0.0902670 | 1.5511684 | -0.0751375 |
| GB45947 | pterin-4-alpha-carbinolamine dehydratase | 98.52058 | 0.1664513 | -0.0523800 | -0.0595421 | -0.0281778 |
| GB46112 | replication factor C subunit 2 | 98.40930 | 0.1725601 | -0.5656149 | -0.1756446 | 0.2166316 |
| GB55662 | mitochondrial fission process protein 1-like | 98.29914 | 0.1521653 | -0.1648426 | 0.0180552 | 0.1917747 |
| GB46775 | bifunctional arginine demethylase and lysyl-hydroxylase PSR isoform X1 | 98.24143 | 0.2934628 | 0.1357369 | 0.7740193 | 0.0693300 |
| GB52799 | methionine aminopeptidase 1D, mitochondrial-like | 98.23116 | -0.1516281 | 0.0732996 | 0.1678067 | 0.2305654 |
| GB52911 | exosome complex component CSL4-like | 98.12694 | 0.3482410 | 0.0411078 | -0.1542678 | 0.1977397 |
| GB55264 | probable serine/threonine-protein kinase DDB_G0283337-like | 98.12283 | -0.1863856 | -0.1572985 | 0.3985734 | -0.0162444 |
| GB45819 | uncharacterized protein LOC725150 | 98.09919 | -0.4450879 | -0.1601616 | 0.5648285 | 0.0373663 |
| GB41392 | heterogeneous nuclear ribonucleoprotein A1, A2/B1 homolog isoform X3 | 98.04981 | 0.0071277 | 0.1151645 | 0.1560302 | -0.0468350 |
| GB51243 | glutathione S-transferase omega-1 | 97.96337 | 0.1249380 | -0.0376622 | 0.0771450 | -0.1212292 |
| GB45684 | protein spire-like isoform X4 | 97.71656 | -0.4064900 | 0.0209486 | 0.2427277 | -0.1816127 |
| GB53647 | WD repeat-containing protein 75-like | 97.64167 | 0.2812419 | 0.0473090 | -0.0133899 | 0.0587657 |
| GB54752 | breast cancer metastasis-suppressor 1-like protein-like isoform 1 | 97.58260 | 0.0552216 | 0.1413598 | -0.0204487 | 0.0642342 |
| GB53821 | telomerase-binding protein EST1A-like isoform X4 | 97.57120 | -0.9237042 | -0.0204516 | -0.5125400 | -0.3198568 |
| GB53131 | acylglycerol kinase, mitochondrial-like | 97.52986 | -0.3515300 | 0.1226544 | -0.1381732 | 0.2512326 |
| GB50772 | uncharacterized protein LOC100578420 isoform X2 | 97.47817 | -0.5310875 | 0.1137321 | -0.1059591 | -0.0439748 |
| GB50272 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase-like isoform X5 | 97.44465 | 0.0460011 | -0.0411525 | -0.1341710 | -0.2369654 |
| 411763 | DNA polymerase subunit gamma-1, mitochondrial | 97.40571 | -0.7385244 | -0.1219991 | 0.0594008 | -0.0821210 |
| GB55091 | translation initiation factor eIF-2B subunit beta | 97.35810 | 0.2003086 | 0.2228942 | -0.2405257 | 0.0239099 |
| GB44689 | E3 ubiquitin-protein ligase synoviolin A-like isoform X3 | 97.32197 | 0.0601999 | 0.0787588 | 0.1062739 | -0.0053641 |
| 100576966 | reticulocyte-binding protein 2 homolog a-like isoform X2 | 97.30328 | -0.4605887 | -0.1083841 | 0.0951272 | 0.1377412 |
| GB48312 | pre-mRNA-splicing factor RBM22-like | 97.30040 | 0.3163381 | -0.0135676 | 0.0938006 | -0.1028333 |
| GB53208 | uncharacterized LOC408620, transcript variant X2 | 97.15218 | 0.1522329 | 0.0893646 | 0.1835043 | -0.0595227 |
| GB46689 | kinesin 4A isoformX1 | 97.13237 | -0.4584644 | -0.0833182 | -0.0603612 | -0.0902299 |
| GB46653 | GPI inositol-deacylase-like isoform X4 | 97.01091 | -0.3391296 | -0.0190985 | -0.0349331 | -0.0234991 |
| GB40590 | metallo-beta-lactamase domain-containing protein 1-like isoform X2 | 96.92196 | 0.2500262 | 0.1342444 | 0.2364377 | 0.1518002 |
| GB18213 | meteorin precursor | 96.88487 | 0.1736949 | 0.1853657 | 0.1710970 | 0.1081279 |
| GB40712 | malectin-like | 96.87963 | 0.0429452 | 0.0108306 | 0.0968702 | -0.2145103 |
| GB55223 | transcription initiation factor TFIID subunit 1 isoform X1 | 96.86710 | 0.2913703 | 0.1980952 | 0.0689959 | -0.0358969 |
| GB46273 | uncharacterized protein LOC409083 | 96.80065 | 0.2862710 | 0.0444399 | -0.2850645 | 0.0782929 |
| GB41472 | DNA-directed RNA polymerase III subunit RPC3-like | 96.77235 | -0.0185853 | -0.0196177 | 0.1974508 | 0.1560408 |
| GB50322 | WD repeat domain phosphoinositide-interacting protein 4-like isoform X2 | 96.74620 | 0.0463944 | -0.0968322 | 0.0146000 | 0.0107335 |
| GB40415 | protein brambleberry-like | 96.61913 | -0.2434614 | 0.1347445 | -0.1932929 | 0.0821312 |
| GB49330 | nicastrin isoform X1 | 96.60803 | 0.1536412 | -0.0370351 | -0.1061660 | 0.0345264 |
| 724533 | small nuclear ribonucleoprotein Sm D3 isoform X1 | 96.60587 | 0.2362631 | 0.1015437 | -0.1600384 | 0.0166429 |
| GB55538 | uncharacterized protein LOC409034 | 96.60289 | -0.4539915 | 0.0812493 | 0.8424342 | -0.0298563 |
| GB51129 | cyclin-dependent kinase 20-like isoform 1 | 96.55732 | -0.6448613 | 0.0291297 | 0.1358084 | -0.1345240 |
| GB55824 | probable serine hydrolase-like isoform X3 | 96.44226 | 0.1986069 | -0.1527330 | -0.0377111 | 0.0360636 |
| GB52029 | eukaryotic translation initiation factor 3 subunit G-like | 96.34038 | 0.5883366 | 0.0062951 | -0.2591365 | 0.0687756 |
| GB47235 | protein UBASH3A homolog | 96.32427 | -0.0849285 | -0.0706748 | 0.4047275 | 0.0155020 |
| GB50552 | oxysterol-binding protein-related protein 11-like | 96.30960 | 0.3267309 | 0.0775224 | 0.3200843 | -0.0058456 |
| GB45045 | uncharacterized protein LOC552753 isoform X3 | 96.26688 | -0.4596840 | 0.1059586 | 0.1166623 | 0.0256063 |
| GB55247 | UDP-N-acetylglucosamine–peptide N-acetylglucosaminyltransferase 110 kDa subunit isoform 2 | 96.25230 | -0.1715209 | -0.0796207 | 0.2598054 | -0.1845775 |
| GB54259 | NAD-dependent protein deacetylase Sirt2 isoform X4 | 96.18695 | 0.2536253 | -0.0321462 | -0.1249548 | 0.0062112 |
| GB43990 | peptidyl-tRNA hydrolase ICT1, mitochondrial-like | 96.15094 | -0.0876257 | 0.0800120 | 0.0312399 | -0.0324967 |
| GB47304 | 5-formyltetrahydrofolate cyclo-ligase-like | 96.14601 | -0.1075994 | 0.0542300 | -0.0237679 | -0.0362518 |
| GB42066 | kinesin 3D isoform X2 | 96.09442 | -0.1180087 | 0.1164174 | -0.0233135 | 0.0875068 |
| GB47191 | uncharacterized protein LOC100576348 isoform X2 | 96.09262 | -0.6475983 | 0.3492833 | -0.3852339 | 0.2046412 |
| GB47105 | nucleolar MIF4G domain-containing protein 1 homolog isoform X1 | 95.94502 | -1.0411937 | 0.1427566 | 0.0546727 | -0.0888035 |
| GB53240 | E3 ubiquitin-protein ligase RNF25-like isoformX1 | 95.75265 | -0.2758530 | 0.0819541 | -0.2312311 | -0.0575227 |
| GB45009 | katanin p80 WD40 repeat-containing subunit B1 isoform X2 | 95.62678 | -0.4636426 | 0.0977194 | -0.2368156 | 0.0835737 |
| 551467 | syntaxin-18 isoform X2 | 95.60963 | -0.4824560 | -0.0989850 | -0.0740157 | 0.1503028 |
| GB44492 | isopentenyl-diphosphate Delta-isomerase 1-like isoformX1 | 95.60887 | -0.4291358 | -1.2839654 | -0.0997690 | 0.0410078 |
| GB50012 | uncharacterized protein LOC726323 isoform X1 | 95.60231 | 0.3273334 | -0.0577196 | 0.1516841 | 0.4737291 |
| GB46218 | transferrin isoform X4 | 95.58149 | 0.0619027 | -0.0640118 | 0.0138558 | -0.2058052 |
| GB52796 | translin | 95.46532 | 0.1722316 | 0.0859329 | -0.0667155 | 0.1746505 |
| GB52198 | LOW QUALITY PROTEIN: actin-binding protein anillin | 95.44314 | 0.1463543 | 0.1008733 | 0.2097859 | -0.0817833 |
| GB54837 | protein MAK16 homolog A-like | 95.36673 | -0.1382445 | 0.4344837 | -0.1597808 | -0.2313941 |
| GB54388 | RNA 3’-terminal phosphate cyclase-like isoform X1 | 95.28097 | 0.3031768 | 0.0651291 | -0.0929415 | 0.1992910 |
| GB47315 | mitochondrial thiamine pyrophosphate carrier-like isoform X2 | 95.18060 | -0.1585082 | -0.0871257 | -0.0072399 | 0.9453269 |
| GB50365 | ski oncogene | 95.17597 | -0.2300198 | -0.0462017 | 0.1810374 | 0.0231927 |
| GB50962 | POU domain protein CF1A-like | 95.16549 | -1.1247133 | -0.1225077 | -0.0992581 | -0.1522642 |
| 552237 | transmembrane protein 242-like | 95.08437 | -0.1791799 | -0.0715258 | 0.0276531 | -0.1270237 |
| GB55485 | DNA methyltransferase 3 | 95.08084 | -0.5874641 | -0.0801435 | -0.0836881 | 0.0236521 |
| GB52744 | poly(A) RNA polymerase, mitochondrial-like isoform X2 | 95.03899 | -0.5308960 | 0.0361782 | -0.0304829 | 0.1779329 |
| GB40446 | unconventional prefoldin RPB5 interactor-like | 95.03779 | -1.4828813 | -0.0034895 | 0.2579053 | 0.1604642 |
| GB53302 | mini-chromosome maintenance complex-binding protein isoform 2 | 94.97034 | -0.2953019 | 0.0077672 | 0.4077361 | 0.0760118 |
| GB42263 | RNA-binding protein 40-like | 94.94164 | -0.4951007 | 0.1600727 | -0.1660684 | 0.3104967 |
| GB44999 | chascon-like | 94.93942 | -0.6330133 | -0.3241300 | 0.2799470 | -0.2112737 |
| GB53246 | protein unc-119 homolog B | 94.86761 | -0.0350373 | 0.0720405 | 0.1201924 | 0.0529067 |
| GB46980 | probable tRNA N6-adenosine threonylcarbamoyltransferase | 94.81051 | 0.1315517 | -0.0117101 | 0.1403145 | -0.0602946 |
| GB51396 | hydroxysteroid dehydrogenase-like protein 2-like isoform X2 | 94.77277 | -0.2374204 | 0.0924130 | -0.0963337 | 0.0246006 |
| GB41295 | nogo-B receptor-like | 94.75637 | 0.0958533 | 0.0380937 | 0.0656854 | -0.0357791 |
| GB53289 | uncharacterized protein LOC552029 isoform X1 | 94.53852 | 0.5070273 | -0.0188373 | -0.5347784 | -0.0068270 |
| GB45378 | thrombospondin-3 isoform X2 | 94.41932 | -0.0770084 | -0.2507295 | -0.2601379 | 0.0172791 |
| GB49574 | uncharacterized protein LOC552171 isoform X1 | 94.37833 | -0.4251797 | -0.0442826 | 0.0451925 | -0.1524273 |
| GB45295 | importin subunit alpha-2-like isoform X1 | 94.34309 | 0.0552061 | 0.1789859 | 0.1930522 | 0.0141187 |
| GB53676 | protein smoothened isoform X2 | 94.30730 | -0.3449464 | 0.0311071 | -0.5157579 | 0.2618153 |
| GB53802 | dipeptidyl peptidase 9-like | 94.26286 | 0.3430208 | 0.1884472 | 0.1642313 | -0.0412584 |
| GB42690 | FAD-linked sulfhydryl oxidase ALR isoformX1 | 94.22280 | 0.2692975 | -0.0042398 | 0.0118296 | 0.1237852 |
| GB40106 | EGF domain-specific O-linked N-acetylglucosamine transferase-like isoform X5 | 94.15941 | 0.1148612 | 0.1102818 | 0.0196236 | -0.0992210 |
| GB42206 | uncharacterized protein LOC100578913 isoform X1 | 94.06032 | -0.7261422 | 0.2704441 | -0.0506108 | 0.0237029 |
| GB41179 | zinc finger protein 569-like | 93.98678 | -0.3936925 | -0.4451640 | 0.1390863 | 0.0165297 |
| GB43121 | uncharacterized protein F21D5.5-like isoform X2 | 93.94919 | -0.1066363 | 0.0882362 | 0.2307165 | 0.4764706 |
| GB53418 | HD domain-containing protein 2-like isoform X1 | 93.94900 | -0.0809322 | -0.2408641 | 0.0755288 | 0.2466224 |
| GB49100 | probable tRNA pseudouridine synthase 2-like | 93.89405 | -0.0249829 | 0.0214858 | 0.0726497 | 0.1857197 |
| GB51541 | protein Hook homolog 3-like isoform 1 | 93.82711 | -0.2802174 | 0.0683751 | 0.0043525 | -0.1468906 |
| GB40336 | neutral ceramidase isoform X1 | 93.82467 | -0.2172815 | -0.0209370 | 1.2435980 | -0.1733590 |
| GB50886 | mediator of RNA polymerase II transcription subunit 30 isoform 1 | 93.76072 | 0.2810430 | 0.0376696 | -0.1308983 | 0.2308189 |
| GB51900 | surfeit locus protein 6 homolog | 93.75773 | -0.6434648 | -0.1066969 | 0.4253944 | 0.3364999 |
| GB41517 | uncharacterized protein LOC100578546 isoform X1 | 93.74356 | -0.7069203 | -0.1630150 | -0.0595612 | 0.0121490 |
| GB49415 | ATP synthase mitochondrial F1 complex assembly factor 2-like | 93.69414 | -0.0314145 | -0.1036498 | -0.0147971 | -0.0961003 |
| GB51394 | poly(ADP-ribose) glycohydrolase ARH3-like | 93.56890 | 0.0603564 | 0.0851963 | 0.0199196 | 0.0958508 |
| GB45680 | UPF0536 protein C12orf66 homolog isoform X2 | 93.56042 | -0.1046661 | 0.0157279 | 0.2772191 | 0.0885859 |
| GB45492 | protein misato-like isoform X1 | 93.51513 | 0.0068212 | -0.1071692 | 0.0694383 | 0.1477929 |
| GB54242 | ribonucleases P/MRP protein subunit POP1-like | 93.48655 | -0.1967686 | 0.1620998 | 0.1363882 | 0.0460275 |
| GB55560 | SH2B adapter protein 1-like isoform X1 | 93.43682 | 0.4433606 | 0.0599952 | 0.1723346 | -0.0489350 |
| GB52503 | integrator complex subunit 7-like isoformX1 | 93.34505 | 0.2150206 | 0.0608692 | 0.0447569 | -0.0775890 |
| GB55837 | MLX-interacting protein isoform X2 | 93.18636 | -0.2540362 | -0.1551088 | -0.0549102 | -0.0446344 |
| 102654737 | peptide deformylase, mitochondrial-like | 93.12917 | -0.2554518 | 0.3849649 | 0.0145548 | 0.1894199 |
| GB51537 | 28S ribosomal protein S11, mitochondrial isoform X1 | 93.06067 | -0.2470972 | 0.0503367 | 0.0554409 | 0.1332603 |
| GB53695 | fatty-acid amide hydrolase 2-like, transcript variant X3 | 93.03614 | 0.6361273 | -0.2265300 | -0.1928881 | 0.0390553 |
| GB44799 | uncharacterized protein LOC552039 isoform X1 | 92.77796 | -0.8875739 | 0.0065999 | -0.0141414 | -0.0415917 |
| GB53716 | estrogen sulfotransferase-like | 92.75099 | -0.0350864 | 0.3435152 | 0.0365727 | 0.1257038 |
| GB44659 | uncharacterized protein LOC100577941 isoform X2 | 92.65827 | -0.3204513 | 0.1152375 | 0.1006300 | 0.1338965 |
| GB51943 | short coiled-coil protein homolog | 92.65275 | 0.2410625 | -0.0102107 | 0.1620607 | 0.2488032 |
| GB44886 | glycine-rich cell wall structural protein 1.8-like isoform X1 | 92.64948 | 0.2095647 | 0.1602760 | 0.1213634 | 0.2425696 |
| GB46501 | tRNA:m(4)X modification enzyme TRM13 homolog | 92.53989 | -0.0594015 | -0.1133658 | 0.0821655 | -0.2085400 |
| GB53387 | GTP-binding protein Di-Ras2-like | 92.49583 | -0.1545741 | -0.0543776 | 0.5089875 | -0.3377624 |
| GB46920 | iron-sulfur cluster assembly enzyme ISCU, mitochondrial | 92.43534 | 0.1073840 | -1.0314564 | 0.1151684 | -0.0992589 |
| GB53861 | mitochondrial tRNA-specific 2-thiouridylase 1-like | 92.34612 | -0.4081187 | -0.1160160 | 0.2351303 | 0.1513819 |
| GB49516 | calcium and integrin-binding protein 1-like isoform X2 | 91.97270 | 0.1561814 | 0.0337540 | 0.2311157 | 0.1315818 |
| GB49953 | vacuolar protein sorting-associated protein 45 isoform 1 | 91.76603 | -0.4801051 | -0.0789026 | 0.0278001 | 0.0544385 |
| GB53532 | serine/threonine-protein kinase STE20-like | 91.60705 | 0.3969424 | -0.0897346 | -0.0625458 | -0.2669483 |
| 102656291 | coiled-coil domain-containing protein 115-like | 91.60297 | -0.0796903 | -0.1916341 | 0.0325521 | -0.1607303 |
| GB40983 | methylosome protein 50 isoform X2 | 91.34296 | 0.3823657 | 0.1548898 | 0.0122354 | -0.0503740 |
| GB50063 | DDB1- and CUL4-associated factor 10-like | 91.31214 | 0.0328908 | 0.1310338 | -0.0057803 | 0.0418550 |
| GB47651 | dnaJ homolog subfamily B member 12-like | 91.28490 | -0.1784141 | -0.0328870 | -0.1689952 | 0.0596196 |
| GB42015 | tubulin polyglutamylase TTLL7-like | 91.23821 | -0.2235526 | 0.0201467 | 0.2891226 | 0.1779291 |
| 100578654 | uncharacterized protein LOC100578654 isoform X4 | 91.11902 | -0.5538979 | 0.1387042 | 0.2300069 | 0.2106901 |
| GB46452 | DNA replication factor Cdt1 isoform X1 | 91.03539 | -0.1345737 | 0.2837372 | -0.0467454 | 0.1549507 |
| GB49629 | ubiquitin carboxyl-terminal hydrolase isoform X2 | 90.93071 | 0.4187357 | -0.0142951 | -0.0752116 | -0.0415293 |
| GB54293 | uncharacterized protein LOC724457 | 90.74656 | -0.3294021 | 0.2137087 | 0.3536797 | 0.1142500 |
| GB55801 | uncharacterized protein LOC413738 isoform X2 | 90.63653 | -0.5194718 | 0.1049738 | -0.2004740 | -0.1322972 |
| GB45688 | synaptic vesicle membrane protein VAT-1 homolog-like | 90.59794 | -0.3034135 | 0.1263921 | 0.1804847 | -0.0632328 |
| GB51635 | centrin-1 | 90.58797 | -0.5322533 | -0.1183215 | 0.0762927 | 0.0077090 |
| GB48944 | vesicle transport protein SEC20-like isoform X1 | 90.56714 | 0.3366751 | 0.0554363 | -0.0034284 | 0.2557447 |
| GB46960 | polypeptide N-acetylgalactosaminyltransferase 5-like isoform X1 | 90.55703 | -0.0948943 | -0.0007394 | 0.6793108 | 0.0334308 |
| GB50941 | phosphatidate phosphatase LPIN2-like isoform X1 | 90.55384 | 0.1207815 | -0.1096204 | 0.0713860 | -0.1255440 |
| GB48677 | protein O-mannosyl-transferase 2-like isoform X2 | 90.53177 | 0.9059295 | 0.1246935 | 0.1772220 | 0.0219852 |
| GB53015 | protein FAM151A-like isoform X3 | 90.38470 | -0.6849796 | 0.2003805 | 0.0631963 | 0.0104523 |
| GB42321 | protein SGT1 homolog ecdysoneless isoform X1 | 90.34536 | -0.2046670 | -0.0392681 | -0.0070138 | -0.0097728 |
| GB41452 | serine/threonine-protein phosphatase PGAM5, mitochondrial-like | 90.27918 | 0.0901027 | 0.3118315 | -0.0645422 | 0.1932577 |
| GB49540 | dynactin subunit 4 | 90.24576 | 0.1646850 | 0.0816541 | 0.1351345 | -0.0242987 |
| GB53847 | abhydrolase domain-containing protein 2 | 90.21311 | 0.1692020 | 0.0116080 | 0.0305612 | 0.0407714 |
| GB50359 | UHRF1-binding protein 1-like isoform X5 | 90.18544 | -0.0397751 | -0.0582086 | 0.0549535 | -0.3414281 |
| GB49352 | transmembrane protein 59-like | 90.08202 | 0.3116883 | 0.0703026 | 0.1377753 | 0.0505254 |
| GB41792 | putative uncharacterized protein DDB_G0282133-like isoform X2 | 90.02222 | -0.0537082 | 0.2942951 | 0.4217642 | 0.0132605 |
| GB50004 | protein preli-like isoform 1 | 90.01359 | -0.3895062 | 0.0249827 | -0.0417402 | 0.0618812 |
| GB46748 | 39S ribosomal protein L54, mitochondrial | 90.00473 | -0.1841175 | 0.0841938 | -0.2336455 | 0.2113185 |
| GB44210 | contactin | 89.79112 | -0.1331464 | -0.0913999 | 0.1158376 | -0.1967150 |
| GB42265 | nuclear envelope phosphatase-regulatory subunit 1-like isoform X2 | 89.74969 | 0.2349445 | 0.0842158 | 0.2057079 | 0.0301283 |
| GB46222 | odorant binding protein 13 precursor | 89.71494 | 0.5543802 | -3.4689759 | 0.1206016 | -0.0265572 |
| GB40717 | B-cell CLL/lymphoma 7 protein family member B-like isoform X1 | 89.68580 | -0.0898894 | 0.2735373 | 0.0632998 | 0.0820293 |
| GB43564 | uncharacterized protein LOC100578443 | 89.67075 | -0.6737872 | 0.3647607 | 0.0514503 | 0.1048131 |
| GB51518 | probable serine/threonine-protein kinase dyrk2-like | 89.37760 | -0.0004133 | 0.0122997 | 0.1444012 | 0.2343744 |
| GB51015 | acylamino-acid-releasing enzyme-like isoform X2 | 89.37110 | 0.0201372 | 0.1366339 | 0.1330789 | 0.0897596 |
| GB52105 | putative uncharacterized protein DDB_G0271606-like isoform X2 | 89.30089 | -0.4411929 | 0.5547986 | -0.1321549 | 0.2895366 |
| GB43246 | uncharacterized protein KIAA0513-like isoform X2 | 89.29393 | -0.8503421 | 0.0924646 | 0.1442277 | 0.1123992 |
| GB45456 | flocculation protein FLO11-like isoform X2 | 89.26424 | -0.1418935 | -0.0530076 | 0.4223401 | -0.1373626 |
| GB48061 | uncharacterized protein LOC413052 | 89.26398 | -0.1461587 | 0.2129896 | -0.2284677 | 0.0256383 |
| GB52998 | radial spoke head protein 9 homolog | 89.19724 | -0.3968545 | -0.6900518 | 0.2269336 | -0.1240472 |
| GB49284 | mucolipin-3 isoform 2 | 89.13228 | 0.1250136 | 0.0341408 | 0.0745202 | 0.0680570 |
| GB55092 | suppressor of fused homolog | 89.12699 | -0.4331377 | 0.1041542 | 0.1457209 | -0.1619385 |
| GB41260 | uncharacterized protein LOC414002 | 89.04329 | 0.1780831 | 0.0012781 | -0.3828629 | 0.9208742 |
| GB40269 | nucleolar protein 14 homolog isoform X1 | 89.00333 | -0.0743241 | -0.0878343 | -0.1234408 | 0.3371177 |
| GB44800 | f-box only protein 33-like, transcript variant X3 | 88.97755 | -0.1172438 | 0.0536991 | 0.0472356 | 0.0333950 |
| GB42086 | single-strand selective monofunctional uracil DNA glycosylase-like | 88.96521 | 0.1991725 | -0.0592079 | 0.2047978 | 0.2161977 |
| 102655420 | uncharacterized protein LOC102655420 | 88.83765 | -0.2653811 | 0.1051354 | 0.3037282 | 0.7730218 |
| GB46741 | growth hormone-regulated TBC protein 1-A isoform X1 | 88.75274 | -0.1828512 | 0.1236310 | 0.1301753 | 0.3032578 |
| GB52652 | uncharacterized protein LOC724152 | 88.67600 | 0.0557587 | 0.2957686 | 0.1100580 | 0.2974843 |
| GB40796 | phospholipase DDHD1-like isoform X3 | 88.64003 | -0.0263786 | -0.0760152 | 0.0165858 | -0.2344648 |
| GB50173 | TATA box-binding protein-associated factor RNA polymerase I subunit B-like | 88.63451 | -0.9366487 | -0.1507002 | 0.0362161 | 0.1542302 |
| GB40067 | dual serine/threonine and tyrosine protein kinase isoform X1 | 88.60116 | -0.1192028 | 0.0979819 | 0.0500681 | -0.0711621 |
| GB47768 | rac GTPase-activating protein 1-like | 88.57621 | -0.0058123 | 0.2453945 | 0.4399285 | 0.3446512 |
| GB43480 | cleavage and polyadenylation specificity factor 160, transcript variant X2 | 88.49929 | 0.2048033 | 0.1463178 | -0.0827978 | -0.0654200 |
| GB51106 | mRNA turnover protein 4 homolog | 88.48936 | 0.3291953 | 0.1048503 | -0.1426921 | 0.2043811 |
| GB55371 | probable asparagine–tRNA ligase, mitochondrial-like isoform X1 | 88.46750 | -0.1009478 | 0.0177979 | 0.0387814 | 0.2122506 |
| GB41829 | ubiquitin-like modifier-activating enzyme atg7-like isoform X3 | 88.32975 | -0.0345944 | -0.0776130 | 0.1496162 | 0.0442789 |
| GB48636 | exosome complex component RRP46 | 88.27582 | 0.2667908 | -0.0346639 | 0.0866776 | 0.0439973 |
| GB40941 | ornithine decarboxylase-like isoform X1 | 88.20129 | 0.3750709 | 1.8748619 | 0.0913002 | 0.9397733 |
| GB43551 | uncharacterized protein LOC409563 | 88.15816 | -0.1190080 | 0.0939626 | -0.0670017 | -0.0596609 |
| GB44054 | DNA replication complex GINS protein PSF1-like | 88.12711 | 0.2513171 | 0.0905309 | -0.0720299 | 0.2465212 |
| GB45830 | kinesin 8 | 87.88068 | -1.6624050 | 0.2796933 | 0.5808391 | 0.0558278 |
| GB50352 | glutathione synthetase isoform X1 | 87.63973 | -0.5080905 | 0.0409502 | 2.1326183 | 0.0862652 |
| GB47992 | splicing factor 3B subunit 4 | 87.54580 | -0.6343011 | 0.1636712 | -0.0344913 | -0.0047916 |
| GB41979 | UTP–glucose-1-phosphate uridylyltransferase isoform X3 | 87.53836 | -0.3227726 | -0.0279324 | 0.0633688 | 0.0664004 |
| GB51421 | CTP synthase, transcript variant X4 | 87.44705 | -0.3568542 | 0.0583338 | -0.0693529 | 0.1393475 |
| GB49449 | uncharacterized protein LOC409179 isoform 1 | 87.43142 | -0.4381497 | -0.8399741 | 0.3667830 | -0.0617186 |
| GB46928 | lys-63-specific deubiquitinase BRCC36-like isoform X2 | 87.40989 | 0.1480342 | 0.1597198 | 0.1955520 | 0.1893948 |
| GB49123 | uncharacterized protein LOC100577638 isoform X1 | 87.38900 | 0.0166608 | 0.1625349 | -0.3324857 | -0.0788806 |
| GB44058 | UDP-glucuronosyltransferase 1-3-like | 87.27745 | -0.0830507 | 0.0611422 | 0.2714085 | 0.2425317 |
| GB45674 | protein aurora borealis | 87.20997 | -0.6332116 | 0.1972691 | 0.1643666 | 0.5962880 |
| GB53826 | transcription factor E2F4 isoform X1 | 87.00013 | 0.4756216 | -0.0275878 | 0.5271656 | -0.0170795 |
| GB55002 | uncharacterized protein LOC100578883 | 86.99800 | -0.2627749 | 0.0351093 | 0.2569920 | 0.0842761 |
| GB40022 | hydroxylysine kinase-like isoform X1 | 86.98188 | 0.5579033 | 0.1740116 | -0.0428017 | 0.0854549 |
| GB44434 | probable glucosamine 6-phosphate N-acetyltransferase-like | 86.98105 | -0.1056350 | 0.0077800 | 0.1270555 | 0.0805236 |
| GB52653 | farnesyl pyrophosphate synthase-like | 86.95563 | -0.5089497 | -1.3586611 | 0.0562782 | 0.1317799 |
| GB55007 | serine protease snake isoform X3 | 86.76174 | 0.0097670 | -0.0807828 | -0.0551113 | 1.0641963 |
| GB49384 | uncharacterized protein LOC725157 | 86.38213 | -0.0163509 | 0.3057350 | 0.9665717 | -0.2015295 |
| GB55527 | uncharacterized protein LOC408649 isoform X2 | 86.33531 | -0.1482014 | -0.1332727 | -0.3426257 | -0.3057878 |
| GB49539 | toys are us | 86.26401 | 0.0005182 | 0.0564093 | 0.2462428 | 0.0461149 |
| GB41894 | uncharacterized protein LOC411277 isoform X28 | 85.97591 | -0.5355215 | 0.2339110 | 0.0951953 | -0.2051621 |
| GB44703 | proteasome activator complex subunit 4-like | 85.82923 | -0.2816499 | -0.1590808 | -0.4150473 | -0.1238055 |
| GB54462 | protein LLP homolog | 85.70284 | -0.3072251 | 0.0408783 | -0.2565174 | -0.0053010 |
| GB46617 | rhythmically expressed gene 2 protein-like isoform X1 | 85.65770 | 0.0119956 | -0.0607103 | 0.1333573 | -0.0614694 |
| 102655268 | WD repeat-containing protein 63-like | 85.60235 | -0.1699526 | -0.0734976 | -0.0823594 | 0.0949795 |
| GB40251 | uncharacterized protein LOC552030 isoform X1 | 85.18945 | 0.2912993 | -0.0073507 | -0.3901558 | 0.0701591 |
| GB49343 | transcriptional repressor protein YY1-like isoform 1 | 85.13815 | 0.3312658 | 0.0307314 | 0.0093500 | -0.0125454 |
| GB55317 | protein lunapark-B-like, transcript variant X2 | 85.09443 | -0.0496236 | 0.1410537 | -0.2352945 | 0.0084366 |
| GB47319 | zinc finger protein Elbow-like isoform X1 | 84.90718 | -0.0123360 | 0.1087597 | -0.0361085 | -0.1376438 |
| GB47460 | cell division cycle protein 20 homolog isoform X1 | 84.83402 | -0.3183535 | 0.1663389 | -0.3446409 | 0.1330472 |
| GB40480 | serine/threonine-protein kinase OSR1-like isoform X5 | 84.82442 | -0.2568944 | 0.0271946 | 0.1904097 | 0.0829648 |
| GB45040 | Krueppel-like factor 10-like isoform X1 | 84.47190 | -0.4999720 | -0.1164206 | 0.1313708 | -0.1121145 |
| GB51863 | flotillin-2 isoform X2 | 84.40611 | -0.3281389 | -0.2045650 | 0.0205942 | -0.0096105 |
| GB45609 | flavin-containing monooxygenase FMO GS-OX-like 4-like | 84.04785 | -0.8054658 | -0.2080427 | -0.4021333 | -0.0568395 |
| GB42054 | sodium/potassium-transporting ATPase subunit alpha isoform X5 | 83.97946 | -0.2885692 | -0.0556927 | 0.1781843 | 0.0932312 |
| GB50877 | tyrosine-protein kinase Dnt isoform X1 | 83.94204 | -0.5189231 | -0.1556333 | 0.2036596 | 0.0935233 |
| GB48118 | negative elongation factor E | 83.78858 | -0.0707260 | 0.1302923 | 0.1511311 | 0.0475081 |
| GB45703 | uncharacterized aarF domain-containing protein kinase 1-like isoform 2 | 83.67506 | -0.0438641 | -0.0966016 | -0.3179459 | -0.1445489 |
| GB53953 | mitochondrial coenzyme A transporter SLC25A42-like isoformX1 | 83.47500 | -0.6876433 | -0.2408674 | 0.1306579 | 0.1308437 |
| GB45913 | protein lethal(2)essential for life-like | 83.41472 | 0.2839773 | 0.4565380 | -0.0489928 | -0.2442535 |
| GB47106 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | 83.40057 | -0.2520387 | -0.0002413 | 0.2750778 | -0.0353279 |
| GB53257 | muscle, skeletal receptor tyrosine protein kinase-like isoform X1 | 83.26680 | 0.1565603 | -0.2002746 | -0.0428387 | 0.0114327 |
| GB51487 | proton-coupled amino acid transporter 4-like | 83.25300 | -0.2686654 | -0.0094929 | 0.0878923 | -0.0191898 |
| GB48234 | tyrosine-protein phosphatase Lar-like | 83.23891 | -0.2364183 | -0.1281370 | -0.9072325 | 0.0931086 |
| GB45046 | cell division control protein 6 homolog | 83.21296 | -0.1434414 | 0.0769154 | -0.5399559 | 0.0190697 |
| GB51937 | hiiragi, transcript variant X4 | 83.14930 | 0.3273538 | -0.2331733 | -0.0189732 | 0.0968262 |
| GB54689 | hippocampus abundant transcript 1 protein-like isoform X6 | 83.13136 | 0.0901529 | 0.0081078 | 0.1082695 | -0.4023399 |
| GB54282 | kinetochore protein NDC80 homolog | 83.12022 | -0.1898754 | 0.0714965 | -0.1853376 | 0.2851134 |
| GB54319 | synaptotagmin 20 isoform X5 | 83.08047 | -0.0133632 | -0.0969320 | 0.1037157 | -0.0670666 |
| GB47468 | integrin alpha-8 isoform X1 | 83.07123 | -0.2510995 | -0.2248056 | 0.0648418 | -0.1559634 |
| 102656381 | ceramide phosphoethanolamine synthase-like | 83.05552 | -0.5814417 | -0.2632366 | 0.0176785 | 0.0552885 |
| GB40535 | 39S ribosomal protein L48, mitochondrial isoform X4 | 83.04377 | -0.4857669 | -0.0779778 | -0.0186406 | 0.0155700 |
| GB49071 | uncharacterized protein LOC725791 | 82.95622 | -0.2820828 | 0.1907288 | -0.1384828 | 0.9511804 |
| 551892 | cytoplasmic protein NCK1 isoform X3 | 82.84500 | 0.1116069 | 0.0266183 | 0.1579394 | -0.0589988 |
| GB43095 | BMP-binding endothelial regulator protein isoform X1 | 82.81707 | -0.6405077 | -0.1575684 | 0.1105583 | 0.1391731 |
| 102656425 | CDGSH iron-sulfur domain-containing protein 3, mitochondrial-like | 82.77827 | 0.0349545 | -0.0505657 | 0.0206878 | 0.1643819 |
| GB46580 | protein wntless-like isoform X1 | 82.76926 | 0.3698836 | 0.1001693 | -0.0364676 | 0.1940352 |
| GB53185 | GPN-loop GTPase 3-like | 82.73138 | 0.0642120 | 0.0112817 | 0.2289060 | 0.1025210 |
| GB46630 | uncharacterized protein LOC100577504 isoform X1 | 82.69068 | -0.5643841 | -0.0455396 | 0.6300647 | 0.0011389 |
| GB40904 | uncharacterized protein LOC725211 | 82.66574 | -0.4457482 | 0.1772111 | 0.0447504 | 0.1167974 |
| GB45195 | protein scribble homolog | 82.55630 | 0.2878775 | -0.0079496 | 0.1623161 | -0.1936888 |
| GB48483 | chaoptin-like isoform X2 | 82.50116 | 0.1440189 | 0.1924029 | 0.0772100 | 0.1981336 |
| GB40963 | putative transcription factor SOX-15 isoform X3 | 82.43319 | 0.1196685 | -0.1972679 | -0.0097669 | -0.4911171 |
| GB51348 | rootletin-like isoform X3 | 82.40007 | -0.5380145 | -0.1181323 | 0.8159832 | -0.1210773 |
| GB42808 | neuroligin 5 | 82.32758 | -1.9548325 | 0.4161654 | -0.3797054 | -0.2839900 |
| GB42555 | uncharacterized protein LOC100577221 | 82.25536 | -0.2162291 | 0.2649999 | 0.0361005 | 0.0831645 |
| GB44044 | intraflagellar transport protein 57 homolog isoform X3 | 82.25237 | 0.4301682 | -0.0381347 | 1.4782182 | -0.0532899 |
| GB48776 | coiled-coil domain-containing protein 6-like | 82.24189 | 0.0638946 | 0.1434135 | -0.0610919 | -0.6573213 |
| GB42110 | slowpoke-binding protein isoform X6 | 82.18032 | 0.2520122 | 0.0617007 | 0.3981189 | 0.2588306 |
| 552443 | N-alpha-acetyltransferase 60-like | 82.15701 | 0.1330272 | 0.1864223 | 0.2124667 | -0.0438390 |
| GB50784 | zinc finger matrin-type protein 5-like | 82.10262 | 0.3849393 | -0.0385070 | 0.1225124 | 0.0488668 |
| GB51513 | UPF0183 protein CG7083-like | 82.05350 | 0.0211656 | 0.1101906 | -0.3676869 | 0.1734152 |
| GB47815 | haloacid dehalogenase-like hydrolase domain-containing protein 2-like | 82.04757 | 0.2335904 | 0.2168662 | 0.5866022 | 0.1500966 |
| GB46467 | uncharacterized protein LOC726694 isoform X2 | 82.02291 | 0.4657472 | 0.1143384 | -0.0003244 | 0.1612604 |
| GB48380 | major facilitator superfamily domain-containing protein 8-like isoform X2 | 81.98490 | 0.0846772 | 0.1259957 | 0.1040438 | -0.1701453 |
| GB48321 | vanin-like protein 1-like | 81.90914 | 0.2360990 | -0.0943761 | 0.1626938 | 0.0598845 |
| GB42192 | metallophosphoesterase domain-containing protein 1-like isoform 1 | 81.82112 | 0.1443089 | 0.0198657 | 0.2660362 | 0.3914187 |
| GB47740 | leucine-rich repeat-containing G-protein coupled receptor 4-like | 81.66683 | -0.5303271 | -0.2760097 | -0.2146710 | 0.3886097 |
| GB40704 | dynein intermediate chain 2, ciliary-like isoform X5 | 81.59501 | -0.7724017 | -0.0421367 | 0.3051828 | 0.1678797 |
| GB42183 | uncharacterized protein LOC100578600 | 81.24792 | -0.2412762 | 0.0054646 | 0.3746364 | -0.2302673 |
| GB45851 | dnaJ homolog subfamily C member 22-like | 81.14522 | 0.0719752 | -0.0035030 | -0.0351223 | -0.2560347 |
| GB53539 | thyroid transcription factor 1-associated protein 26 homolog | 81.06653 | 0.0780399 | 0.1355708 | 0.0225715 | 0.0663746 |
| GB43729 | rac guanine nucleotide exchange factor JJ | 81.05302 | -0.0465988 | 0.4973586 | 0.1165652 | 0.3257298 |
| GB54190 | uncharacterized aarF domain-containing protein kinase 4 isoform X4 | 80.86515 | -0.1460776 | -0.0576026 | 0.0695965 | 0.1147598 |
| GB54421 | uncharacterized protein DDB_G0287625-like | 80.68366 | 0.2555447 | -0.0223220 | 0.0225622 | 0.0343484 |
| GB44117 | roundabout homolog 2 isoform X3 | 80.62255 | -0.2439432 | -0.1580854 | 0.5526838 | -0.2752797 |
| 724756 | uncharacterized protein LOC724756 | 80.55652 | 0.0252041 | -0.7245791 | 0.0601137 | 0.0430655 |
| GB54653 | U1 small nuclear ribonucleoprotein A | 80.55328 | 0.4538914 | -0.0564165 | 0.0005411 | 0.1692871 |
| GB55296 | pyroglutamyl-peptidase 1-like | 80.51840 | 0.5804078 | 0.0743039 | 0.1908893 | -0.0825618 |
| GB43159 | NF-X1-type zinc finger protein NFXL1-like | 80.36205 | -0.1483047 | 0.1208877 | -0.2430825 | -0.0368309 |
| GB45139 | uncharacterized protein LOC726903 | 80.31203 | -1.3366787 | 0.5602430 | 0.1349894 | 0.0272964 |
| GB52666 | putative uncharacterized protein DDB_G0282133-like | 79.96871 | -1.0598835 | -0.4212401 | 0.5123482 | -0.1547904 |
| GB52601 | AT-rich interactive domain-containing protein 5B-like isoform X3 | 79.81946 | 0.0633386 | 0.1674699 | 0.0688821 | 0.0286104 |
| GB42231 | beta-1,4-N-acetylgalactosaminyltransferase bre-4 isoform X3 | 79.70121 | -0.3728579 | 0.0089446 | -0.1231586 | 0.2542434 |
| GB54758 | WD repeat, SAM and U-box domain-containing protein 1-like isoform X1 | 79.65627 | 0.0424168 | -0.2816129 | -0.1669858 | -0.0086753 |
| GB44387 | B-cell lymphoma/leukemia 11B-like isoform X3 | 79.54794 | -0.5453242 | 0.1424302 | 0.4329808 | -0.3405438 |
| GB55755 | protein outspread | 79.45071 | -0.0861913 | 0.1010403 | 0.1728972 | 0.0608626 |
| GB42526 | malate dehydrogenase, mitochondrial-like isoform 1 | 79.38029 | -0.2474192 | -0.2335988 | 0.0266447 | 0.0804945 |
| GB40770 | dehydrogenase/reductase SDR family member 11-like isoform X2 | 79.18283 | 0.0327204 | 0.3536748 | 0.3865886 | 0.1673807 |
| GB43220 | transcription termination factor 2 isoform X1 | 79.10265 | 0.2644874 | 0.2759698 | 0.0538725 | -0.1434828 |
| GB53378 | zinc finger protein 28 homolog | 79.02376 | -0.5786499 | -0.1548530 | 0.0113102 | -0.1577209 |
| GB52797 | enhancer of polycomb homolog 1 | 78.89984 | 0.1779685 | 0.0484715 | -0.0194867 | 0.1231170 |
| GB45255 | aurora kinase B isoform X1 | 78.87479 | 0.1692136 | 0.1481645 | -0.1537423 | 0.1794915 |
| GB47938 | uncharacterized protein LOC412825 isoform X1 | 78.84925 | -0.4208665 | -0.0042892 | 0.7579974 | -0.2479282 |
| GB44203 | arrestin domain-containing protein 3 | 78.79010 | 0.2496367 | 0.2087420 | 0.4262637 | 0.0779627 |
| GB45714 | transglutaminase | 78.60881 | -0.3969663 | -0.0184445 | 0.0964747 | -0.0299562 |
| GB50669 | uncharacterized protein LOC410428 | 78.54674 | -0.0373480 | -0.7215476 | 0.1106084 | -0.2177330 |
| GB51714 | stimulator of interferon genes protein-like | 78.44488 | 0.0188718 | -0.0227268 | 0.0198118 | -0.0256844 |
| GB47159 | caspase-1-like | 78.36622 | -0.2970784 | 0.0663003 | 0.0945799 | -0.1894921 |
| GB52077 | period circadian protein | 78.26096 | -0.1280252 | 0.1157650 | -1.1662534 | 0.6476625 |
| GB43261 | ubiquitin thioesterase trabid isoform X1 | 78.20541 | -0.4480047 | 0.0923657 | -0.0073038 | 0.1350528 |
| GB44781 | exocyst complex component 4-like | 78.17615 | 0.4207412 | 0.0228108 | 0.1991691 | 0.2717861 |
| GB41417 | leucine-rich repeats and immunoglobulin-like domains protein 1-like | 78.12317 | -0.2802413 | 0.0220012 | 0.1681992 | -0.3004220 |
| GB49988 | SRR1-like protein-like isoform X2 | 78.05574 | -0.7996147 | 0.0063959 | 0.0828932 | 0.1191617 |
| GB44443 | transmembrane protein 179-like isoform X1 | 78.04143 | -0.0315907 | 0.0167050 | 0.0580752 | 0.0058616 |
| GB47331 | programmed cell death protein 5-like | 78.04013 | 0.1977021 | 0.0515153 | 0.2636433 | 0.1043132 |
| GB54851 | zinc transporter 1-like isoform X7 | 77.94763 | 0.0947167 | 0.2326054 | 0.4317043 | 0.1249878 |
| GB45058 | MIP18 family protein CG7949-like | 77.88119 | 0.1350506 | 0.2720550 | 0.1353205 | 0.3067768 |
| GB40750 | putative ATP-dependent RNA helicase me31b-like isoform 1 | 77.61807 | -0.1704060 | 0.1652610 | 0.0847445 | -0.0768376 |
| GB54158 | intraflagellar transport protein 140 homolog | 77.56391 | 0.1830641 | 0.0752116 | 0.1407870 | -0.2210602 |
| GB45420 | graves disease carrier protein homolog | 77.45713 | -0.1360487 | 0.1423748 | 0.1213185 | 0.0040754 |
| GB40312 | choline/ethanolamine kinase-like isoform X4 | 77.27476 | -0.4635767 | -0.2391946 | 0.0300019 | -0.1293016 |
| GB51242 | uncharacterized protein LOC727370 isoform X3 | 77.25346 | -0.1357094 | -0.0283603 | 0.3313378 | -0.2787618 |
| GB49870 | long-chain-fatty-acid–CoA ligase 6-like isoform X3 | 77.09863 | 0.1810343 | -0.0752888 | 0.0408349 | -0.2176815 |
| GB46145 | sodium/potassium-transporting ATPase subunit beta-2-like | 77.04330 | 0.2699432 | -0.1832867 | 0.1724996 | -0.0408796 |
| GB53531 | A disintegrin and metalloproteinase with thrombospondin motifs 7-like isoform X9 | 76.93675 | -0.8798427 | 0.3755786 | 0.0342842 | -0.0944177 |
| GB42500 | peptidoglycan-recognition protein LC isoform X2 | 76.79466 | 0.3371163 | -0.1100098 | 0.4807124 | 0.0370728 |
| GB52154 | protein prenyltransferase alpha subunit repeat-containing protein 1-like isoform X1 | 76.78576 | 0.3918293 | 0.0190154 | -0.3236200 | 0.1186753 |
| GB45052 | LOW QUALITY PROTEIN: ral guanine nucleotide dissociation stimulator | 76.77655 | 0.0007175 | 0.1210512 | 0.1206847 | -0.1450082 |
| GB46038 | elongation of very long chain fatty acids protein 4-like isoform X2 | 76.74328 | -0.1373967 | 0.3267714 | 0.2502768 | -0.1202751 |
| GB43831 | ATP-binding cassette sub-family D member 3-like | 76.66762 | 0.0779268 | -0.3410303 | -0.1571787 | 0.0660232 |
| GB41921 | adenylate cyclase type 5-like | 76.26866 | -0.0704037 | -0.1245870 | 0.2382754 | 0.0372682 |
| GB54100 | ragulator complex protein LAMTOR4 homolog isoform X2 | 76.13231 | 0.2996614 | 0.0127978 | 0.1548202 | 0.0594167 |
| GB50016 | cell division cycle-associated protein 7-like | 76.13021 | -0.1436496 | 0.2677806 | -0.0802188 | 0.2221760 |
| GB44328 | dynein light chain 1, axonemal-like isoform X2 | 76.10849 | 0.0981617 | -1.1004849 | 0.0396446 | 0.0257270 |
| GB52882 | calcium-independent phospholipase A2-gamma-like isoform X1 | 76.02784 | -0.6250468 | -0.2402948 | 0.0874514 | 0.0770572 |
| GB48310 | zinc transporter ZIP1-like isoform X2 | 75.83697 | -0.2838646 | -0.2170711 | 0.1074896 | -0.0326357 |
| GB42276 | uncharacterized protein LOC550958 | 75.79765 | -0.3924328 | 0.0490164 | 0.2829247 | 0.0531546 |
| GB43427 | homer protein homolog 2-like isoform X2 | 75.65467 | -0.1316778 | 0.0231139 | 0.0587408 | 0.4091429 |
| GB45106 | run domain Beclin-1 interacting and cysteine-rich containing protein-like isoform X1 | 75.60987 | -0.2486185 | 0.0329622 | 0.1058623 | -0.2555359 |
| GB46056 | heart- and neural crest derivatives-expressed protein 1-like isoform X1 | 75.47002 | -0.2891780 | -0.0576471 | 0.2999764 | -0.0424345 |
| GB50933 | GATA-binding factor A | 75.27293 | 0.4701499 | 0.1713163 | 0.1857035 | 0.1884606 |
| GB46490 | rho guanine nucleotide exchange factor 10-like isoform X3 | 75.25832 | -0.1544410 | -0.0197056 | -0.0535951 | 0.1051275 |
| GB49469 | lymphocyte cytosolic protein 2-like isoform X1 | 75.09182 | -0.6202643 | 0.0734105 | -0.0499513 | 0.1704506 |
| GB46245 | integrator complex subunit 12 isoform X5 | 75.06811 | -0.1796624 | 0.0187300 | -0.2108451 | 0.1505376 |
| GB55192 | sorting nexin-16-like | 75.06433 | 0.3271364 | -0.0918411 | 0.0937238 | 0.3094774 |
| GB53518 | 28S ribosomal protein S30, mitochondrial | 75.01202 | -0.3100083 | 1.4938757 | -0.2760613 | 0.2340060 |
| GB50282 | glycine receptor subunit alpha-2 isoform X8 | 74.98614 | -0.0919255 | -0.1228171 | 0.2543653 | 0.1474961 |
| GB53662 | protein HID1-like isoformX1 | 74.84256 | 0.3060871 | 0.1354545 | -0.2192123 | 0.1887126 |
| GB55604 | uncharacterized protein LOC725144 isoform X4 | 74.63560 | 0.1536292 | -0.3071154 | 0.1386568 | 0.4853530 |
| GB44440 | FERM domain-containing protein 8 isoform 1 | 74.57709 | 0.3940632 | -0.0726472 | 0.1882671 | 0.0247747 |
| GB44337 | DNA polymerase delta small subunit isoform X2 | 74.55218 | -0.2086525 | 0.0055189 | -0.2809085 | 0.1501402 |
| 413046 | sarcolemmal membrane-associated protein-like | 74.47378 | -0.2565446 | 0.0329929 | 0.0268377 | -0.3897590 |
| GB52073 | probable citrate synthase 1, mitochondrial-like | 74.38047 | -0.4023298 | -0.1588605 | -0.0628810 | 0.1885859 |
| GB48829 | blood vessel epicardial substance-like, transcript variant X4 | 74.35871 | 0.1896171 | -0.0712744 | 0.0563397 | 0.0232820 |
| GB42719 | E3 ubiquitin-protein ligase MARCH8-like isoform X1 | 74.11818 | 0.3485036 | -0.0921535 | 0.2507317 | 0.0607427 |
| GB52025 | neprilysin 2 isoform X4 | 74.07443 | 0.3741702 | -0.1377266 | 0.2515321 | 0.0582520 |
| GB46395 | 40S ribosomal protein S12, mitochondrial | 74.03483 | 0.3373619 | -0.0544390 | 0.1722078 | 0.0598111 |
| GB42478 | CD2-associated protein | 73.86326 | -0.0005831 | 0.0698841 | 0.1739929 | -0.0826623 |
| GB50290 | uncharacterized protein LOC724917 isoform X2 | 73.84131 | 0.2263371 | 0.3499708 | 0.2695718 | -0.0350391 |
| GB49084 | dual specificity protein kinase TTK-like | 73.75643 | 0.4391332 | 0.1772004 | -0.3232440 | 0.0302207 |
| GB49769 | putative tRNA (cytidine(32)/guanosine(34)-2’-O)-methyltransferase-like isoform 1 | 73.57879 | 0.3505131 | -0.0229773 | -0.8812585 | 0.3794693 |
| 724643 | borealin-like | 73.26823 | 0.0835274 | -0.0693634 | -0.0539582 | -0.0574988 |
| GB51504 | solute carrier family 23 member 1-like | 73.11624 | 0.0835258 | 0.3747462 | 0.0918719 | -0.1470971 |
| GB51118 | 28S ribosomal protein S28, mitochondrial | 73.09820 | 0.0297621 | 0.0819829 | -0.5428747 | 0.0015057 |
| GB51739 | alpha-ketoglutarate-dependent dioxygenase alkB homolog 4-like | 73.03627 | 0.2744241 | 0.1467053 | 0.0545301 | -0.2248307 |
| GB55746 | myosin-IB isoform X2 | 72.92993 | 0.0128099 | -0.0518167 | -0.0272041 | -0.2939229 |
| GB42156 | enhancer of filamentation 1 | 72.90725 | -0.5293556 | 0.1365882 | -0.2005914 | -0.0974010 |
| GB41773 | proteoglycan 4-like isoform X1 | 72.90297 | 0.2517214 | -0.6911315 | 0.1915830 | 0.1824917 |
| GB50978 | general transcription factor IIE subunit 1 | 72.88375 | 0.5505829 | -0.0008111 | 0.0661839 | 0.1300035 |
| GB50435 | zinc finger and BTB domain-containing protein 20-like | 72.48761 | -0.6793546 | 0.0732153 | 0.3980137 | 0.2278010 |
| GB46368 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)]-like | 72.34977 | 0.0298679 | -0.4406704 | -0.2348546 | -0.3117601 |
| 725196 | putative peptidyl-tRNA hydrolase PTRHD1-like isoformX2 | 72.05179 | 0.3978826 | -0.0016375 | 0.3028199 | 0.0498457 |
| GB51558 | dentin sialophosphoprotein-like isoform X2 | 71.89535 | -0.3974676 | -0.1135625 | -0.2204494 | 0.0636279 |
| GB48836 | uncharacterized protein LOC100577578 isoform X1 | 71.83474 | 0.1288726 | 0.1039358 | 0.0723859 | -0.0156149 |
| GB50460 | ras association domain-containing protein 2 | 71.78631 | 0.4585587 | 0.5016901 | 0.0470089 | 0.1202778 |
| GB55840 | protein sidekick-1-like isoform X5 | 71.58552 | 0.1847896 | 0.0070244 | -0.0582451 | -0.0416547 |
| GB41486 | putative neutral sphingomyelinase-like isoform X1 | 71.45629 | -0.0033464 | -0.0528102 | 0.0978353 | 0.0876926 |
| GB50407 | uncharacterized protein LOC100578579 isoform X2 | 71.06894 | -0.7437220 | 0.1885016 | 1.0069762 | 0.2613802 |
| GB51409 | homeobox protein Nkx-6.1-like | 70.82281 | -0.9111943 | -0.9564960 | 0.2917807 | 0.2140245 |
| GB54525 | ras-like protein family member 11B-like | 70.73745 | 0.0476575 | -0.1214252 | 0.1008147 | -0.1737334 |
| GB46537 | cell death-related nuclease 6 isoform X1 | 70.64259 | 0.0845914 | -0.0773749 | 0.0483848 | 0.2116229 |
| GB55909 | uncharacterized protein LOC725992 isoform X1 | 70.56496 | -0.3152749 | -0.1171764 | 0.8843745 | -0.0457715 |
| 102656840 | probable cyclin-dependent serine/threonine-protein kinase DDB_G0292550-like | 70.50464 | -0.4002282 | 0.2248767 | 0.1535262 | 0.0438474 |
| GB42872 | G1/S-specific cyclin-E | 70.25337 | 0.1914332 | 0.3032163 | -0.0240269 | 0.2118319 |
| GB53943 | tRNA methyltransferase 112 homolog | 70.18889 | 0.0370066 | 0.1885109 | -0.2325104 | 0.1581564 |
| GB45525 | kin of IRRE-like protein 3-like isoform X2 | 70.04210 | -0.9642374 | -0.1422867 | 0.1615934 | 0.0941893 |
| 100577213 | tumor necrosis factor receptor superfamily member 10B-like | 70.01077 | -1.1452914 | -0.1691454 | 0.5108142 | -0.1780156 |
| GB50990 | protein FAM76A-like isoform X3 | 69.28323 | 0.3229110 | 0.0016474 | 0.1488574 | 0.0778880 |
| GB45815 | SCY1-like protein 2-like isoform X2 | 69.10924 | -0.0667885 | -0.0596507 | -0.4399345 | 0.4220746 |
| GB40781 | fibroblast growth factor receptor substrate 2-like | 68.90474 | -0.3171705 | 0.2331021 | 0.7553513 | 0.0572574 |
| GB47057 | protein tramtrack, alpha isoform-like isoform X7 | 68.79220 | -0.2612772 | 0.0269970 | 0.0671450 | 0.3998210 |
| GB51817 | glucose dehydrogenase [FAD, quinone] | 68.69944 | -2.5637785 | 0.7384892 | 0.3225832 | 0.1556621 |
| GB43825 | lysosomal aspartic protease | 68.47974 | 0.6021461 | 0.0937831 | 0.8615854 | 1.8961223 |
| GB40871 | protein bric-a-brac 1-like | 68.31938 | -0.4667468 | 0.0246127 | 1.0435273 | 0.0889583 |
| GB55299 | serine/threonine-protein kinase PAK mbt isoform X3 | 68.09970 | -0.1574919 | 0.1371840 | 0.0997319 | 0.0408209 |
| GB41414 | titin-like isoform X3 | 67.97853 | 0.2227736 | -0.3134406 | 0.7665292 | 0.4024970 |
| GB47470 | cadherin-related tumor suppressor | 67.92032 | -1.2828778 | 0.0599174 | 0.6093695 | -0.6214463 |
| GB45841 | ras guanine nucleotide exchange factor P-like isoform X2 | 67.56666 | 0.2774367 | -0.0055298 | 0.0624082 | -0.0653708 |
| GB54522 | Bardet-Biedl syndrome 2 protein homolog isoform X2 | 67.19257 | 0.1878212 | -0.1241067 | 0.3689639 | -0.0461029 |
| GB51515 | uncharacterized LOC100577174, transcript variant X4 | 66.92524 | 0.0487011 | 0.1238639 | 0.0958852 | -0.0929412 |
| GB44462 | cytochrome c oxidase subunit 5A, mitochondrial | 66.92109 | -0.3791897 | -0.2157607 | 0.6898045 | 0.1789873 |
| GB52039 | hairy/enhancer-of-split related with YRPW motif protein 1-like isoform X4 | 66.83893 | -0.7086989 | -0.1028304 | 0.2043076 | -0.0668303 |
| GB50050 | ADP-ribosylation factor-like protein 3-like | 66.79205 | -0.0761433 | -0.1824594 | 0.0229471 | 0.1313307 |
| GB52504 | lachesin-like isoform X4 | 66.46035 | -0.4024399 | 0.4698165 | 0.0810441 | -0.3807297 |
| GB49115 | probable low affinity copper uptake protein 2-like isoform X1 | 66.20512 | -0.2712830 | -0.1177837 | -0.3492182 | 0.1590764 |
| GB53788 | tyrosine-protein phosphatase non-receptor type 9-like isoform 1 | 66.09907 | 0.0665039 | 0.1117453 | 0.4406215 | 0.2292932 |
| GB47804 | peptidoglycan-recognition protein LB isoform X2 | 65.95727 | 0.0098779 | -0.0007363 | 0.1696490 | 0.0098228 |
| GB55486 | histone lysine demethylase PHF8-like isoform X3 | 65.60089 | 0.1421111 | -0.0346205 | 0.1656952 | 0.0461724 |
| GB51014 | EF-hand domain-containing protein CG10641-like isoform X2 | 65.28955 | 0.3613154 | 0.1724833 | -0.0378858 | -0.0103540 |
| GB51418 | uncharacterized protein LOC725844 | 65.26256 | 0.1521675 | -0.1600204 | 0.8616428 | 0.0348054 |
| GB54359 | uncharacterized protein LOC726251 isoform X1 | 65.24457 | 0.8501316 | -0.1421378 | -0.0264114 | -0.0572257 |
| GB42894 | disheveled-associated activator of morphogenesis 1 isoform X1 | 65.08864 | 0.0387441 | -0.1185047 | 0.4577864 | 0.1485406 |
| GB42330 | protein DPCD-like | 64.81360 | 0.2873032 | 0.1494385 | -0.0708865 | 0.1086516 |
| GB54886 | LOW QUALITY PROTEIN: protein GDAP2 homolog | 64.72424 | 0.2352555 | -0.0471316 | 0.0839623 | -0.4307745 |
| 102655356 | thyrotropin-releasing hormone receptor-like | 64.71649 | 0.4707057 | 0.3582825 | 0.2926145 | 0.3059052 |
| 102656439 | adenosine deaminase CECR1-like | 64.65076 | -0.1502085 | -0.0463265 | 1.2832897 | -0.2414131 |
| GB18327 | pyrokinin-like receptor 2 | 64.49841 | -0.7115854 | -0.3042653 | 0.0385518 | -0.1267117 |
| GB52279 | neurexin 1 precursor | 64.30439 | -0.9448179 | 0.2203311 | 0.4762512 | 0.4204253 |
| GB45547 | uncharacterized protein LOC726958 | 64.29820 | 0.2933015 | -0.0472358 | 0.2176309 | 0.2077648 |
| GB45870 | uncharacterized protein LOC551818 | 63.89730 | -0.1902776 | 0.0532255 | 0.3610019 | 0.1820232 |
| GB55531 | rab11 family-interacting protein 1 isoform X2 | 63.84537 | 0.2097737 | -0.0089849 | 0.0953808 | -0.0312325 |
| 102655904 | DNA-directed RNA polymerases I, II, and III subunit RPABC3-like isoform X6 | 63.81509 | 0.3877567 | -0.0725608 | 0.2238133 | -0.0798492 |
| GB50181 | RCC1 domain-containing protein DDB_G0279253-like | 63.56678 | 0.1525277 | -0.0392366 | 0.0951083 | -0.1963366 |
| GB50525 | serine/arginine repetitive matrix protein 2-like isoform X3 | 63.48967 | -0.2321031 | -0.1337802 | 0.1990983 | -0.1474399 |
| GB47185 | uncharacterized protein LOC551433 | 63.48035 | -0.9903041 | -0.9239763 | 0.2812161 | 0.0699502 |
| GB47838 | flocculation protein FLO11-like | 63.33045 | -0.6459520 | -0.3086387 | 1.0126782 | -0.3385289 |
| GB44072 | E3 ubiquitin-protein ligase TRIM9 isoform X2 | 63.24766 | -0.7105547 | 0.1197700 | 0.3339203 | -0.1632030 |
| GB42062 | uncharacterized protein LOC724563 | 63.01870 | -0.0159379 | -0.2795829 | 0.2090740 | 0.2098259 |
| GB40876 | pleckstrin homology domain-containing family J member 1-like | 62.95027 | -0.3288025 | 0.1302610 | -0.0356572 | 0.1944279 |
| GB44045 | protein croquemort isoform X6 | 62.79329 | -0.2163677 | 0.0159170 | -0.2992615 | 0.0330013 |
| GB41975 | cyclin-Y | 62.61946 | -0.2421144 | 0.0363458 | -0.0614363 | 0.1393621 |
| GB45875 | G-protein coupled receptor Mth2-like isoform X2 | 62.43391 | 0.3817644 | -0.1509732 | 0.8212698 | 0.3126977 |
| 724832 | innexin inx2 | 61.70746 | 0.1517293 | 0.1934897 | -0.3190546 | 0.1470327 |
| GB46541 | B(0,+)-type amino acid transporter 1-like isoform X2 | 61.50690 | 0.0683294 | -0.0225892 | 0.2508287 | -0.1600118 |
| GB49738 | photoreceptor-specific nuclear receptor isoform X1 | 60.92910 | 0.2406627 | -0.2892613 | 0.1619275 | -0.2571818 |
| GB52417 | spatacsin, transcript variant X2 | 60.76413 | -0.2159015 | -0.0688343 | 1.1431620 | 0.0780870 |
| GB49655 | uncharacterized protein LOC726100 isoform X1 | 60.68510 | -0.0398938 | -0.0207831 | -0.0073220 | 0.2094088 |
| GB43909 | plasma membrane calcium-transporting ATPase 3 isoform X1 | 60.42327 | -0.0717766 | -0.0683675 | 0.0439869 | -0.0410479 |
| GB42412 | tubulin delta chain-like | 59.98401 | -0.8131011 | -0.1104493 | 0.0222230 | 0.0381230 |
| GB49807 | cell cycle checkpoint protein RAD1 | 59.59164 | -0.1794221 | 0.0652831 | 0.1917067 | 0.2196132 |
| GB50620 | peptidyl-prolyl cis-trans isomerase-like | 59.18989 | 0.0163374 | 0.0346210 | -0.1198109 | 0.2204073 |
| GB43130 | single Ig IL-1-related receptor-like | 58.93308 | 0.1697364 | 0.2185858 | -0.0188302 | -0.2928417 |
| GB50648 | serine proteinase stubble isoform X1 | 58.74000 | -0.9247339 | -0.0887801 | 0.3230406 | 0.4094308 |
| GB52947 | protein mesh-like isoform X2 | 58.65355 | 0.6160398 | 0.1705780 | 0.1355985 | 0.2805696 |
| GB41734 | reversion-inducing-cysteine-rich protein with kazal motifs isoform X2 | 58.58734 | -0.5098348 | 0.0544886 | 0.2156190 | 0.1059428 |
| 102656594 | uncharacterized protein LOC102656594 | 58.46945 | 0.1010416 | 0.0056006 | 0.3214435 | 0.0087650 |
| GB46060 | UPF0501 protein KIAA1430 homolog | 58.30671 | 0.0252131 | 0.1278140 | 0.2153479 | -0.1359132 |
| GB41591 | putative glucose-6-phosphate 1-epimerase-like | 57.82898 | 0.2733866 | 0.1734465 | 0.1066729 | 0.2132363 |
| GB46376 | protein LSM14 homolog A isoform X1 | 57.77768 | -0.0359011 | 0.2511789 | 0.0581416 | 0.2918125 |
| GB51736 | tweedle motif cuticular protein 2 | 57.41328 | -0.1708556 | 0.3323590 | 0.1818311 | 0.4852333 |
| GB42758 | forkhead box protein D3-like | 57.21339 | -0.2353991 | -0.0105576 | 0.0915022 | -0.1879695 |
| GB40799 | protein HOS4-like isoform X1 | 57.07600 | -0.0929235 | 0.0652953 | 0.4530388 | 0.1594375 |
| GB54778 | CAD protein isoform 1 | 56.94917 | -0.1316537 | 0.1638002 | 0.8838453 | -0.3362347 |
| GB44464 | ubiquinone biosynthesis protein COQ7 | 56.73898 | 0.2628560 | 0.0480824 | 0.1787419 | -0.1173611 |
| GB52756 | apyrase precursor | 56.41373 | -0.0832512 | -0.1085466 | 0.0584467 | 0.0602014 |
| GB53625 | uncharacterized protein LOC411622 | 56.37878 | 0.1395023 | -0.2898437 | 0.1560606 | 0.0324178 |
| GB45073 | fibrillin-2-like | 55.38762 | -0.2637609 | -0.3453368 | 0.7762066 | -0.6774509 |
| GB51613 | uncharacterized protein LOC408570 isoform X1 | 55.33685 | 0.5553386 | -0.1551220 | 1.0905932 | 0.5852404 |
| GB54268 | nicotinate phosphoribosyltransferase-like isoform X3 | 55.13134 | 0.0151011 | -0.0255891 | -0.0575809 | 0.0197632 |
| GB49416 | protein msta, isoform A-like isoform X3 | 54.01849 | 0.0159441 | -0.1873364 | 0.5657615 | -0.5931037 |
| GB46398 | thyrotroph embryonic factor isoformX1 | 53.62674 | -0.9310904 | 0.2375295 | -0.0979614 | -0.0104330 |
| GB55516 | bone morphogenetic protein 2-B isoform X2 | 53.33755 | -0.0057689 | 0.0401956 | 0.2185907 | 0.5014711 |
| GB51331 | tektin-4-like | 53.26269 | 0.2398246 | -0.1448660 | -0.0114410 | 0.1740892 |
| GB45403 | innexin inx1-like | 53.22721 | 0.0057850 | 0.1700522 | 0.2551215 | 0.0874559 |
| GB47970 | alpha-aminoadipic semialdehyde synthase, mitochondrial | 53.22618 | 0.0036304 | -0.0962658 | 0.0137247 | 0.0340244 |
| GB52700 | uncharacterized protein LOC410520 | 53.18054 | -1.5603852 | -0.0460095 | -0.0763453 | -1.0769759 |
| GB54499 | molybdenum cofactor sulfurase-like isoform X3 | 53.06893 | 0.0290549 | -0.0719749 | 0.0700159 | 0.1118500 |
| GB51938 | leucine rich repeat G protein coupled receptor | 53.01662 | -0.4037629 | 0.5053746 | 0.1885290 | 0.0499168 |
| GB49862 | uncharacterized protein LOC724773 isoform X1 | 52.85923 | -0.5242188 | -0.2540650 | 0.1607607 | -0.2321009 |
| GB50257 | uncharacterized protein LOC408508 isoform X1 | 52.80735 | -0.0362760 | 0.1809520 | -0.4082392 | 0.2420318 |
| GB50129 | tektin-3-like | 52.58387 | -1.0760862 | -0.3458620 | 1.3452251 | 0.0724621 |
| 102656337 | mitotic spindle assembly checkpoint protein MAD2A-like isoform X1 | 52.34531 | 0.2786206 | 0.1718437 | -0.0561645 | 0.3612307 |
| 102653588 | cleavage and polyadenylation specificity factor 73-like | 51.69641 | -0.1388123 | -0.0529202 | 1.9604224 | -0.1036728 |
| GB43158 | nuclear pore complex protein Nup205 | 51.15607 | 0.0531488 | 0.1495736 | 0.8006445 | 0.0622148 |
| GB51560 | histone-lysine N-methyltransferase 2D-like isoform X1 | 51.14060 | 0.5870860 | 1.1297587 | 0.5931453 | -0.0850856 |
| GB54295 | beta-1-syntrophin isoform X3 | 51.07129 | -0.5961330 | 0.2034047 | -0.8783025 | -0.4991127 |
| GB50745 | sphingosine kinase 2-like isoform X3 | 51.02808 | 0.0362962 | -0.1725146 | 0.2085656 | -0.1847709 |
| GB47849 | pyrroline-5-carboxylate reductase 2-like isoform X2 | 50.91214 | -0.0079302 | -0.2430339 | 0.0248888 | 0.6111611 |
| GB40437 | adenomatous polyposis coli protein-like | 50.52433 | -0.4570451 | 0.0825313 | -0.0379912 | -0.1944207 |
| GB55505 | open rectifier potassium channel protein 1-like | 50.45895 | -0.1804009 | -0.1825428 | 0.1016832 | -0.0514651 |
| GB52791 | ammonium transporter 1-like | 50.26889 | -0.6510191 | -0.0709328 | 0.2307484 | 0.2822120 |
| 100578205 | uncharacterized protein LOC100578205 | 50.23934 | -0.9996975 | 0.1679468 | -0.2736439 | 0.5058984 |
| GB48271 | broad-complex isoform X10 | 50.19891 | -0.3264123 | -0.9120557 | 0.3661601 | 0.3676537 |
| GB42812 | uncharacterized SDCCAG3 family protein-like | 49.48408 | -0.0537222 | -0.1026573 | -1.0757927 | 0.0419329 |
| GB52614 | REST corepressor 3 isoformX2 | 49.11374 | 0.4039927 | 0.1530866 | 0.1275605 | 0.1361707 |
| GB43877 | aquaporin AQPcic-like isoform X2 | 48.97815 | 0.2743879 | 0.1263906 | 0.3627980 | -0.0080933 |
| GB48543 | uncharacterized protein LOC100577936 isoform X5 | 48.83182 | -0.4903594 | -0.2120898 | -0.1008715 | -0.2597354 |
| GB41027 | BAG family molecular chaperone regulator 2-like | 48.74667 | 0.2521315 | 0.0389659 | -0.3967533 | 0.0917787 |
| GB41583 | cysteine-rich PDZ-binding protein | 48.72062 | 0.1588802 | 0.0457086 | 0.1889442 | 0.3023709 |
| GB41296 | uncharacterized protein LOC100578542 precursor | 48.66540 | 0.1464229 | -0.3591324 | 1.7522270 | 0.2166730 |
| GB54239 | zinc finger protein 853 isoform X6 | 48.40169 | 0.3000663 | -0.0687687 | -0.0055985 | 0.0456838 |
| 100577273 | uncharacterized protein LOC100577273 | 48.19120 | 0.1279529 | 0.7481239 | 0.1511708 | -0.2342699 |
| GB44163 | transmembrane and TPR repeat-containing protein CG4341-like isoform X1 | 47.99393 | 0.1886519 | 0.0920604 | 0.1520100 | 0.1803761 |
| GB41760 | lipase 3-like | 47.86088 | 0.1987229 | 0.4916942 | 0.3682522 | 0.3250953 |
| GB48086 | uncharacterized protein LOC551512 | 47.61673 | 0.1452494 | 0.0096495 | 0.0682472 | 0.0422986 |
| GB49149 | neurogenic locus Notch protein isoform X3 | 47.48812 | -0.2222969 | 0.2137294 | -0.0012329 | -0.1195830 |
| GB41115 | Kv channel-interacting protein 1-like isoform X5 | 47.39684 | -1.0616212 | 0.1832864 | 0.3516676 | -0.4092668 |
| 102655819 | extensin-like | 47.27870 | -0.4510156 | -0.9163967 | 0.9484551 | -0.2868919 |
| GB44043 | uncharacterized protein LOC724216 | 47.14662 | 0.5168608 | -1.3710471 | 0.6925880 | -0.5326878 |
| GB44829 | bifunctional purine biosynthesis protein PURH-like isoform X2 | 46.74025 | -0.0006758 | -0.0501562 | 0.4026800 | 0.0861745 |
| GB48446 | interferon regulatory factor 2-binding protein-like B-like | 46.68582 | -0.0672429 | 0.0519734 | 0.2015959 | -0.9135980 |
| GB44170 | intraflagellar transport protein 43 homolog isoform X4 | 45.89960 | 0.1172748 | 0.0179837 | 0.5435114 | -0.1708747 |
| GB55834 | uncharacterized protein LOC726282 | 45.59171 | 0.1360734 | 0.4479348 | 0.7575685 | -0.0757016 |
| GB41182 | proton-associated sugar transporter A-like isoform 1 | 45.27011 | 0.4659433 | -0.0092487 | 0.0615205 | -0.2746701 |
| GB50113 | mitogen-activated protein kinase kinase kinase 15-like isoform X3 | 43.84136 | 0.6085096 | -0.0682533 | 0.1691411 | 0.2290563 |
| 102656841 | WD repeat-containing protein WRAP73-like | 43.10688 | -0.4081065 | 0.0389685 | -0.5288194 | 0.0535406 |
| GB50094 | MMS19 nucleotide excision repair protein homolog | 42.87412 | -0.0697374 | -0.2026910 | 0.2241561 | -0.3217248 |
| 102655306 | uncharacterized protein LOC102655306 | 41.89924 | -0.8361760 | 0.4347175 | 0.4053602 | -0.0115265 |
| GB46663 | uncharacterized protein LOC410375 isoform X2 | 41.85294 | 0.1848931 | -0.0531352 | 0.0263710 | 0.0658579 |
| GB50402 | uncharacterized protein LOC412801 | 41.66312 | -0.3061277 | -0.2847435 | 0.5059297 | -0.3866755 |
| GB46296 | slit homolog 1 protein-like | 41.11175 | -0.0344174 | 0.0044020 | -0.1650275 | 0.0422088 |
| GB48694 | probable inactive protein kinase DDB_G0270444-like isoform X2 | 41.00674 | -0.9867388 | 0.1988047 | 0.2814439 | 0.2802956 |
| GB44577 | solute carrier family 35 member G1-like | 40.87708 | -0.3001515 | -0.0023463 | 0.8842170 | NA |
| GB47635 | suppressor of variegation 3-9 isoform X2 | 40.78175 | 0.1472413 | 0.0664006 | 0.2667078 | 0.0347691 |
| GB47199 | bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial isoform X2 | 40.66767 | -0.0412714 | 0.0275652 | 0.1725019 | -0.2279684 |
| GB49794 | uncharacterized protein LOC100577530 isoform X3 | 39.72344 | -0.2210553 | 0.0912312 | 0.2545806 | -0.0680889 |
| GB54817 | muscle-specific protein 20 | 39.53068 | 0.5054273 | -0.1022903 | 0.3433915 | 0.0893511 |
| GB47579 | vesicular glutamate transporter 3 isoform X2 | 39.00617 | -0.0788585 | -0.2609606 | 0.1051467 | -0.0942900 |
| GB52517 | oxysterol-binding protein-related protein 1-like isoformX1 | 38.94730 | -0.0648616 | 0.0327672 | 0.3061130 | 0.0098350 |
| GB54765 | cytochrome P450 18a1 | 35.95987 | -1.0594604 | 0.0586940 | 0.0430433 | -0.5771207 |
| GB44783 | putative GTP cyclohydrolase 1 type 2 Nif3l1-like | 35.90006 | 0.3311548 | 0.0178708 | 0.0138595 | 1.0805774 |
| GB50763 | arrestin domain-containing protein 2 | 35.76708 | -0.1285922 | 0.0395234 | 0.1553251 | -0.7556396 |
| GB19642 | troponin C type IIa | 35.62903 | 0.4253569 | -0.5991469 | 0.2855267 | 0.1694526 |
| GB43860 | protein ELYS-like isoform X3 | 35.55157 | -0.3521961 | 0.0626613 | 0.4992968 | -0.1149052 |
| GB51013 | T-related protein-like isoform X2 | 35.36915 | 0.2661110 | -0.3048160 | 0.3234008 | -0.7060438 |
| GB45344 | probable phosphatase phospho2-like | 35.31529 | 0.5535069 | -0.0146228 | 0.1994171 | -0.1196679 |
| GB13325 | chemosensory protein 6 precursor | 34.92767 | 0.8386459 | -0.0081968 | 0.1835364 | 0.7685853 |
| GB49079 | estradiol 17-beta-dehydrogenase 8-like | 33.90588 | 0.5773511 | -0.0851843 | -0.4223868 | 0.0102120 |
| GB50845 | uncharacterized protein LOC725891 isoform X5 | 33.63126 | 1.1653787 | -0.3772206 | 0.4313499 | 0.2741969 |
| GB46437 | histone chaperone asf1 | 33.55454 | -0.2510559 | 0.2098430 | 0.2038942 | 0.0779987 |
| GB46286 | zinc carboxypeptidase A 1-like isoform X1 | 33.38593 | 0.4642732 | 0.3880605 | 0.4536903 | -0.2305250 |
| GB50062 | sulfotransferase 1C4-like | 33.34935 | -0.4984718 | -0.7360999 | 0.1208885 | 1.2023152 |
| GB45076 | transcription factor Sox-21-B-like | 32.98055 | -1.4364426 | 0.8753540 | 0.6971990 | -0.5555090 |
| GB48028 | circadian locomoter output cycles protein kaput | 32.81373 | 0.0999370 | -0.1207097 | -0.2512987 | 0.1248088 |
| GB42300 | uncharacterized protein LOC100577920 | 32.66700 | -0.6277404 | 0.0516783 | 0.2900979 | 0.2462532 |
| GB48270 | uncharacterized protein LOC100577045 isoform X1 | 31.61988 | 0.6296575 | -0.0395377 | 0.3347949 | 0.1779517 |
| GB44913 | fringe glycosyltransferase isoform X1 | 31.44783 | 0.6965961 | 0.0329426 | 0.1261872 | 0.2355285 |
| GB49541 | zinc finger SWIM domain-containing protein 8-like isoform X3 | 31.36059 | -0.0863004 | 0.1482970 | -0.3694119 | 0.0822621 |
| GB47805 | peptidoglycan-recognition protein S2 isoform X1 | 30.43869 | 0.5130847 | 0.0102949 | -0.0542260 | 0.3830162 |
| GB41695 | transcription factor Sox-10-like isoform X1 | 30.15929 | -0.1617476 | -0.2052570 | 0.0879113 | -0.4831547 |
| GB51292 | homeobox protein H90 | 29.95961 | -1.5717484 | 0.2605334 | -0.2242671 | 0.0479805 |
| GB52040 | popeye domain-containing protein 3-like | 28.47698 | -0.6500925 | -0.2791416 | 0.2676382 | 0.0385765 |
| GB53025 | sister chromatid cohesion protein DCC1-like | 24.61047 | -0.2549911 | 0.2814634 | -0.1452519 | 0.3348280 |
| 102655272 | NHP2-like protein 1-like | 23.81786 | 0.7631749 | 0.1487093 | -1.6188682 | 0.0676269 |
Module 2
Table S27: List of all the genes in Module 2, ranked by their within-module connectivity, \(k\). The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
gene_list <- module_gene_list(2)
saveRDS(gene_list, file = "supplement/tab_S27.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB43105 | casein kinase II subunit alpha isoform X6 | 82.779541 | 0.3899105 | 0.0790337 | 0.0091448 | 0.0169172 |
| GB40946 | serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform-like isoform X1 | 80.538588 | 0.4085220 | 0.0719860 | 0.0454678 | 0.0178463 |
| GB45257 | ubiquitin-conjugating enzyme E2 L3-like isoform 2 | 80.094381 | 0.3617777 | -0.0299000 | -0.0057910 | 0.1267934 |
| GB53180 | importin subunit alpha-3 | 79.075096 | 0.5678582 | 0.0364811 | -0.0212055 | -0.0225912 |
| GB53723 | cytoplasmic tRNA 2-thiolation protein 1-like | 78.714595 | 0.0514981 | 0.0446512 | 0.0289127 | 0.0386278 |
| GB43750 | prefoldin subunit 5-like | 77.478531 | 0.5561579 | 0.1149251 | -0.0003891 | 0.0341238 |
| GB43742 | thioredoxin 1-like 1 isoform 1 | 77.173530 | 0.4209603 | -0.0210857 | 0.2473120 | -0.0689485 |
| GB51414 | mRNA export factor-like | 76.222672 | 0.4047987 | 0.0893679 | -0.1083921 | 0.0576379 |
| GB40429 | ras-related protein Rab-11A isoform X1 | 75.954541 | 0.2469198 | -0.0608410 | -0.3090996 | 0.0560599 |
| GB46322 | electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial isoform X2 | 74.438935 | 0.3642614 | -0.1334926 | -0.1350087 | 0.1225978 |
| GB45181 | probable Bax inhibitor 1 | 74.401365 | 0.4470905 | -0.0666689 | -0.1669489 | -0.2705991 |
| GB53311 | neuralized-like protein 2-like | 72.979406 | 0.7559364 | -0.0231900 | 0.5787808 | 0.0139633 |
| GB55232 | 3-hydroxyacyl-CoA dehydrogenase type-2-like | 72.680153 | 0.6767801 | 0.0350239 | -0.1210798 | 0.2163465 |
| GB45127 | transmembrane 9 superfamily member 4-like isoform 1 | 72.448290 | 0.2315142 | -0.0807298 | -0.1055090 | -0.0041899 |
| GB51710 | eukaryotic initiation factor 4A-like isoformX2 | 72.193983 | 0.3309664 | -0.0062636 | 0.2603149 | 0.3071035 |
| GB50369 | ankyrin repeat and FYVE domain-containing protein 1-like isoform X3 | 71.979643 | 0.2708422 | 0.0356164 | -0.7052720 | 0.0465993 |
| GB56034 | nuclear migration protein nudC-like | 71.171411 | 0.5010790 | 0.0665033 | 0.0234851 | 0.0770466 |
| GB45752 | ubiquitin-conjugating enzyme E2 N | 69.697025 | 0.4236299 | 0.1418321 | 0.0016739 | 0.5385159 |
| GB49192 | ribosomal RNA small subunit methyltransferase NEP1-like | 69.453530 | 0.3097282 | 0.0144822 | -0.1003551 | 0.0004354 |
| GB55434 | rab GDP dissociation inhibitor beta | 68.379381 | 0.3532497 | 0.0506273 | -0.1635366 | -0.3187517 |
| GB50080 | calmodulin-like protein 4-like | 68.210793 | 0.6356881 | 0.0852815 | -0.0993701 | -0.0554956 |
| GB48487 | xylosyltransferase oxt | 68.192030 | 0.3098182 | -0.0137376 | -0.0434064 | 0.0144619 |
| 552579 | proteasome subunit beta type-3-like | 67.350603 | 0.4186865 | -0.0716817 | -0.1044372 | 0.0315463 |
| GB44693 | complement component 1 Q subcomponent-binding protein, mitochondrial-like | 67.275201 | 0.0634121 | 0.1181245 | -0.0278215 | 0.0880520 |
| GB51009 | T-complex protein 1 subunit delta-like isoform 1 | 66.897641 | 0.6491909 | 0.0219451 | 0.0734191 | -0.0577987 |
| GB44312 | hydroxyacylglutathione hydrolase, mitochondrial-like isoform X2 | 66.474024 | 0.4281245 | -0.0354899 | -0.0453722 | 0.0527255 |
| GB48369 | lysM and putative peptidoglycan-binding domain-containing protein 1-like isoform X2 | 66.443097 | 0.3609862 | -0.0933002 | -0.0076084 | 0.1088149 |
| GB53382 | cytosolic Fe-S cluster assembly factor NUBP1 homolog isoform X1 | 66.352637 | 0.4559343 | -0.0619590 | -0.0181799 | 0.0431696 |
| GB47478 | glutathione peroxidase-like 1 | 65.944626 | 0.2031135 | -0.0122454 | -0.2598539 | -0.3733075 |
| GB51333 | coatomer subunit gamma isoform X3 | 65.732174 | 0.4601223 | 0.0981151 | -0.0742245 | -0.0280520 |
| GB50731 | ragulator complex protein LAMTOR1-like | 65.449433 | 0.5385336 | 0.1081764 | 0.8160667 | -0.0406650 |
| GB51065 | 40S ribosomal protein S10-like isoform 1 | 65.323225 | 0.6008137 | 0.0628584 | -0.2291556 | -0.0091458 |
| GB45285 | eukaryotic translation initiation factor 3 subunit F-like | 65.166123 | 0.5837586 | 0.0345384 | 0.0428350 | -0.0349192 |
| GB53349 | proteasome subunit alpha type-3 | 64.904659 | 0.3093979 | -0.0735537 | -0.1904000 | -0.1817445 |
| GB49597 | eukaryotic translation initiation factor 4B-like | 64.753478 | 0.4181901 | 0.0397658 | -0.0731381 | 0.1006943 |
| GB55568 | short-chain specific acyl-CoA dehydrogenase, mitochondrial isoform X1 | 64.411735 | 0.4896179 | -0.0348562 | 0.0069892 | -0.1003224 |
| GB55572 | S-phase kinase-associated protein 1 isoform 1 | 64.297278 | 0.1502337 | 0.0103900 | -0.3256861 | 0.0396846 |
| GB45047 | proteasome subunit beta type-7-like | 64.215028 | 0.3781791 | -0.0678040 | 0.4018756 | -0.0769933 |
| GB45354 | V-type proton ATPase subunit d isoform X1 | 63.727478 | 0.3323942 | -0.0317595 | 0.0216173 | -0.0028670 |
| GB44576 | ester hydrolase C11orf54 homolog | 63.638054 | 0.1890940 | -0.0014018 | 0.2807424 | 0.2776325 |
| GB48072 | eukaryotic translation initiation factor 3 subunit M | 63.521086 | 0.3070557 | 0.0892132 | 0.0210727 | 0.0951780 |
| GB50598 | aldose reductase-like isoform 1 | 63.496085 | 0.5400739 | 0.2243442 | -0.0377091 | -0.2016640 |
| GB51072 | 40S ribosomal protein S4-like isoform 1 | 62.457789 | 0.6691693 | 0.0755220 | -0.1558421 | -0.1441905 |
| GB48916 | charged multivesicular body protein 6-like | 62.202785 | 0.2544770 | 0.0432777 | -0.1154126 | -0.0473375 |
| GB48669 | neutral and basic amino acid transport protein rBAT isoform X3 | 62.107824 | 0.2328783 | -0.0290059 | 0.0726677 | 0.0858647 |
| GB41358 | elongation factor 1-alpha | 61.731114 | 0.8151329 | 0.0700644 | -0.2097850 | NA |
| GB51497 | DNA-directed RNA polymerase II subunit RPB11 | 61.629369 | 0.2523410 | -0.0415946 | 0.0144893 | 0.0995764 |
| GB41664 | group XIIA secretory phospholipase A2-like isoform X1 | 61.328010 | 0.2658127 | -0.0302407 | 0.0560136 | 0.0996414 |
| GB44755 | eukaryotic translation initiation factor 4E type 3-A-like | 61.144935 | 0.1038005 | 0.1238230 | 0.0373720 | -0.6723524 |
| GB55625 | NEDD4 family-interacting protein 1-like | 60.731413 | 0.5654293 | -0.0125876 | -0.0039550 | 0.0090174 |
| GB43147 | proteasome subunit beta type-2-like isoform 1 | 60.664909 | 0.3079746 | -0.1349309 | -0.0459174 | 0.0588287 |
| GB40653 | 60S ribosomal protein L24 | 60.471702 | 0.5381179 | 0.0425044 | -0.1804111 | 0.1994310 |
| GB54784 | ATP-dependent RNA helicase DDX42-like isoform X2 | 60.315962 | 0.2414669 | 0.0847043 | -0.0247280 | 0.0133743 |
| GB47655 | protein AAR2 homolog | 60.035350 | 0.3305521 | 0.1761220 | -0.0186990 | -0.0660279 |
| GB55420 | programmed cell death protein 6-like isoform 3 | 59.941234 | 0.5828077 | -0.0819480 | 0.0200568 | 0.8212542 |
| GB43852 | ubiquitin-like protein 4A-like isoform 2 | 59.610870 | 0.3051302 | 0.0387012 | -0.0238347 | 0.1142672 |
| GB40539 | 40S ribosomal protein S20 | 59.556613 | 0.5181401 | 0.0718535 | -0.1277454 | -0.0451962 |
| GB55639 | 40S ribosomal protein S3 | 58.751362 | 0.6660898 | 0.0144752 | 0.1362449 | -0.0812502 |
| GB50057 | dihydroorotate dehydrogenase (quinone), mitochondrial-like | 58.648425 | 0.1933239 | -0.0531178 | 0.1113681 | 0.1511399 |
| GB41363 | 26S protease regulatory subunit 6B isoform 1 | 58.459081 | 0.5410512 | -0.0136308 | -0.0270127 | 0.0203711 |
| GB47998 | AP-2 complex subunit mu isoform 1 | 58.347165 | 0.3301556 | -0.0335771 | -0.0048470 | -0.5143606 |
| GB42560 | 14-3-3 protein epsilon isoform X2 | 58.339471 | 0.5044700 | 0.0369770 | 0.1695536 | -0.0780955 |
| GB53626 | myotrophin-like isoform 2 | 58.008840 | 0.5073563 | 0.1610072 | -0.0604751 | 0.1382949 |
| GB54379 | cleavage stimulation factor subunit 1-like | 57.944057 | 0.5957286 | 0.1892855 | 0.0290971 | 0.0109216 |
| GB44704 | flavin reductase (NADPH)-like | 57.916538 | 0.2107071 | 0.0867890 | 0.0238738 | -0.0130074 |
| GB46420 | eukaryotic translation initiation factor 3 subunit I isoform X1 | 57.072059 | 0.5630588 | 0.1272195 | -0.0568560 | 0.0321002 |
| GB45018 | prolactin regulatory element-binding protein-like | 56.995307 | 0.3388228 | 0.0271635 | -0.0186523 | -0.0721073 |
| GB45880 | ubiquinone biosynthesis protein COQ4 homolog, mitochondrial-like isoform X3 | 56.946969 | 0.2255158 | -0.0648247 | 0.0143367 | 0.0134140 |
| GB50333 | 40S ribosomal protein S6-like | 56.907873 | 0.3803377 | 0.2920983 | -0.0166515 | 0.0469258 |
| 410306 | ADP-ribosylation factor 2-like | 56.596623 | 0.3596849 | 0.0122742 | -0.0693314 | -0.0121140 |
| GB54211 | protein DJ-1-like | 56.344516 | 0.3136343 | 0.0014124 | 0.7698125 | 0.0166411 |
| GB40576 | 60S acidic ribosomal protein P0 isoform X2 | 56.090846 | 0.7419921 | 0.1036318 | -0.0189775 | 0.1186527 |
| GB41211 | ATP-binding cassette sub-family E member 1 | 55.949884 | 0.7874174 | 0.0996597 | -0.0588728 | -0.0078862 |
| GB40414 | protein SMG8-like | 55.864277 | 0.1311333 | 0.0260062 | 0.3267312 | -0.0158654 |
| GB48810 | 60S ribosomal protein L8 | 55.819919 | 0.5374155 | 0.0864496 | 0.0594938 | 0.0886045 |
| GB18750 | T-cell immunomodulatory protein isoform X1 | 55.698119 | 0.4093780 | 0.0152509 | -0.0393054 | -0.0955995 |
| GB42679 | 40S ribosomal protein S8 | 55.513452 | 0.2985759 | 0.0726029 | -0.1494389 | 0.1072492 |
| GB52728 | prohibitin-2-like | 55.364673 | 0.2622934 | 0.0968903 | 0.0467469 | 0.0321292 |
| GB54797 | brahma-associated protein of 60 kDa-like isoform X2 | 55.360103 | 0.3609989 | 0.0245313 | -0.1375910 | 0.0428848 |
| GB46540 | cytochrome b5-like isoform X2 | 55.297320 | 0.4561460 | 0.1260684 | -0.1412306 | 0.0714888 |
| GB45171 | SUMO-conjugating enzyme UBC9 isoform X2 | 55.126919 | 0.5777018 | -0.0071439 | -0.0952634 | 0.0741116 |
| GB47103 | elongation factor 1-beta’ | 55.076449 | 0.3100352 | 0.0947439 | -0.0233222 | 0.2622463 |
| GB52116 | 60S ribosomal protein L36 isoform X2 | 54.840113 | 0.5609103 | 0.0633467 | 0.0034544 | -0.0399395 |
| GB42537 | 40S ribosomal protein S15 | 54.821238 | 0.7335142 | 0.0675785 | -0.0002162 | 0.0342294 |
| GB54747 | 26S proteasome non-ATPase regulatory subunit 11-like | 54.722684 | 0.3825784 | -0.0310609 | -0.1317055 | -0.0194236 |
| GB54973 | selT-like protein-like isoform 1 | 54.677274 | 0.6929597 | -0.0817881 | 0.0294361 | -0.0229722 |
| GB43392 | guanine deaminase-like | 54.428508 | 0.2595250 | -0.0557265 | 0.2379932 | -0.6192207 |
| GB54814 | 60S ribosomal protein L31 isoform 1 | 54.369542 | 0.7211279 | 0.0815471 | -0.0702305 | -0.1517580 |
| GB40718 | thioredoxin reductase 1 isoform X1 | 54.110820 | 0.3976980 | -0.0923669 | 0.1908874 | 0.0141603 |
| GB46888 | alpha-methylacyl-CoA racemase-like | 53.922636 | 0.4389072 | -0.1826352 | -0.6123075 | -0.1119572 |
| GB55901 | ribosome biogenesis protein NSA2 homolog isoform X1 | 53.877207 | 0.4878904 | -0.0036286 | -0.0417069 | -0.0789324 |
| GB53402 | calcineurin B homologous protein 1 | 53.641579 | 0.3779003 | -0.1525626 | -0.1477300 | -0.1176049 |
| GB45856 | protein GPR107-like isoform X4 | 53.484790 | 0.4014230 | -0.0503593 | -0.1783136 | 0.0155807 |
| GB53849 | segment polarity protein dishevelled homolog DVL-3 isoform X2 | 53.479444 | 0.4988536 | 0.0988280 | 0.0008910 | 0.8151443 |
| GB47925 | coatomer subunit beta | 53.402838 | 0.5520184 | 0.1110889 | -0.0674825 | 0.3316757 |
| GB52694 | proteasome subunit beta type-4-like | 53.270162 | 0.5807396 | -0.0642302 | -0.1116811 | -0.0137662 |
| GB49377 | 40S ribosomal protein S3a | 53.267539 | 0.5013839 | 0.0272403 | 0.2853204 | 0.0019517 |
| GB44997 | palmitoyltransferase ZDHHC5-like isoform X1 | 52.976965 | 0.1054200 | 0.1607361 | 0.0125694 | 0.0378256 |
| GB50652 | T-complex protein 1 subunit epsilon | 52.915369 | 0.7727276 | 0.1218119 | -0.4867803 | 0.0174350 |
| GB53360 | V-type proton ATPase subunit D 1-like isoform 1 | 52.782785 | 0.1923389 | -0.0209061 | 0.0346521 | 0.6134235 |
| GB51033 | E3 ubiquitin-protein ligase parkin-like isoform 1 | 52.691442 | 0.2712819 | -0.0369253 | -0.0357602 | 0.0192391 |
| GB51683 | annexin-B9-like isoform X1 | 52.637726 | 0.5436988 | -0.0376760 | 0.0689681 | -0.0325108 |
| GB50158 | 60S ribosomal protein L4 isoform 1 | 52.599917 | 0.7484971 | 0.1027183 | 0.1668092 | 0.1127432 |
| GB55011 | eukaryotic translation initiation factor 3 subunit K-like | 52.504836 | 0.3517346 | 0.0330282 | -0.0599971 | 0.0271320 |
| GB44749 | 60S ribosomal protein L9 | 52.408203 | 0.6183303 | 0.0584793 | -0.1931285 | 0.1710115 |
| GB42036 | protein SEC13 homolog isoform X2 | 52.387806 | 0.4850137 | 0.0510745 | 0.0181188 | 0.8574785 |
| GB45261 | mannose-1-phosphate guanyltransferase beta-like | 52.324926 | 0.4964924 | 0.0599660 | 0.0055369 | -0.0066053 |
| GB47880 | superoxide dismutase 1 | 52.265335 | 0.4669031 | -0.3461375 | -0.0193634 | -0.3238591 |
| GB40341 | uncharacterized protein LOC413618 | 52.059747 | 0.1733059 | 0.0828853 | -0.0880786 | 0.0376459 |
| GB54183 | DNA-directed RNA polymerases I, II, and III subunit RPABC2-like | 51.823221 | 0.2400619 | 0.0464102 | -0.1435838 | 0.1383333 |
| GB44160 | protein dpy-30 homolog | 51.691222 | 0.6329524 | 0.1202659 | -0.1786552 | -0.0467911 |
| GB45369 | receptor of activated protein kinase C 1, transcript variant X3 | 51.674572 | 0.6061490 | 0.0987082 | 0.0099143 | -0.1744317 |
| GB50870 | 39S ribosomal protein L41, mitochondrial | 51.531271 | 0.0882601 | 0.0205089 | -0.0402980 | 0.0514872 |
| GB43559 | 60S ribosomal protein L3 | 51.530487 | 0.6010300 | 0.1141575 | 0.0020165 | 0.1404949 |
| GB50519 | transmembrane emp24 domain-containing protein eca-like | 51.316906 | 0.7096674 | 0.0156954 | 0.3864642 | -0.8619462 |
| GB48536 | T-complex protein 1 subunit beta-like isoform 1 | 51.251431 | 0.7461775 | 0.0972661 | 0.0278174 | -0.0214331 |
| GB51889 | vesicle-trafficking protein SEC22b-B-like isoform X1 | 51.132352 | 0.4296799 | -0.0444747 | -0.0715219 | 0.0663911 |
| GB53974 | probable RNA helicase armi | 51.115561 | 0.1423549 | 0.2440794 | 0.5106117 | 0.1041671 |
| GB54854 | proteasome maturation protein-like | 51.069422 | 0.5228479 | -0.0802985 | -0.0483899 | -0.0077136 |
| GB50455 | ubiquitin-conjugating enzyme E2-17 kDa-like | 51.033981 | 0.6761680 | 0.0520493 | 0.0526095 | 0.2928392 |
| GB53799 | proteasome subunit alpha type-2 | 50.979772 | 0.6359607 | -0.0960932 | -0.1479950 | -0.0181309 |
| GB42152 | puromycin-sensitive aminopeptidase isoform X3 | 50.954882 | 0.1569441 | 0.0437223 | -0.0293714 | -0.0880934 |
| GB42810 | uncharacterized protein C7orf26 homolog | 50.806485 | 0.4203041 | -0.0287058 | -0.0536175 | 0.0291954 |
| GB50917 | 60S acidic ribosomal protein P1 | 50.680174 | 0.6271819 | 0.0677499 | 0.0325577 | 0.1420895 |
| GB43466 | retinol dehydrogenase 11-like | 50.633644 | 0.3269587 | -0.0443370 | -0.0360959 | 0.1183058 |
| GB51973 | methionine aminopeptidase 1-like | 50.380731 | 0.2620277 | -0.0378810 | 0.0823959 | 0.0843747 |
| GB54779 | syntenin-1-like isoform X1 | 50.289759 | 0.4066501 | -0.0324619 | 0.0160917 | -0.0574747 |
| GB47638 | ER membrane protein complex subunit 3-like | 50.175741 | 0.7047659 | -0.0103128 | -0.0468210 | 0.0482700 |
| GB46750 | 40S ribosomal protein S16 | 50.099142 | 0.6200634 | 0.0900704 | 0.5508549 | -0.4241530 |
| GB45737 | target of rapamycin complex subunit lst8-like | 50.031832 | 0.4024227 | 0.1178256 | 0.0377706 | 0.1993536 |
| GB52512 | 60S ribosomal protein L28 | 50.019615 | 0.7142262 | 0.0644402 | 0.3955537 | -0.0487567 |
| GB48150 | actin-related protein 2/3 complex subunit 1A | 49.842642 | 0.5254966 | 0.0383029 | -0.0457504 | -0.0498008 |
| GB55748 | exosome complex component RRP40 | 49.785262 | 0.3223877 | 0.1683391 | -0.1277459 | 0.0471182 |
| GB53219 | 40S ribosomal protein S17 | 49.704548 | 0.7418752 | 0.0967735 | 0.1160953 | -0.2496817 |
| 409728 | 40S ribosomal protein S5 isoform X1 | 49.449695 | 0.9166193 | 0.0687048 | 0.1743880 | 0.1204288 |
| GB51201 | 40S ribosomal protein S12 isoform X1 | 49.398062 | 0.7241416 | 0.0665665 | -0.0115462 | 0.0278723 |
| GB44039 | malate dehydrogenase, cytoplasmic-like isoform 1 | 49.341750 | 0.5132430 | -0.1385158 | -0.0072436 | 0.1747652 |
| GB55077 | ras suppressor protein 1 isoform X2 | 49.163900 | 0.2799454 | -0.1094379 | 0.0624915 | 0.0908027 |
| GB46562 | 40S ribosomal protein S24-like isoform X2 | 49.143478 | 0.3207560 | 0.0310005 | 0.1096683 | 0.0507754 |
| 410017 | protein OPI10 homolog | 49.120658 | 0.5637709 | 0.0736794 | -0.2682441 | 0.0110299 |
| GB46776 | 40S ribosomal protein S11 isoform X1 | 49.040441 | 0.6238829 | 0.0884087 | -0.0580724 | -0.2446433 |
| GB48309 | uncharacterized protein LOC552534 | 48.952661 | 0.2012036 | 0.0296439 | 0.0455195 | 0.0495045 |
| GB51038 | 60S ribosomal protein L23 | 48.907138 | 0.7391449 | 0.0806656 | -0.1850061 | -0.0270500 |
| GB40232 | peroxiredoxin 1 | 48.840666 | 0.7901203 | 0.1037701 | 0.4673685 | 0.2752115 |
| GB41604 | chloride intracellular channel exc-4 | 48.699728 | 0.2227495 | -0.0207292 | 0.2289713 | 0.2574692 |
| GB50356 | 60S acidic ribosomal protein P2 | 48.671273 | 0.8365958 | 0.0997759 | -0.0280343 | -0.0520760 |
| GB53321 | transmembrane protein adipocyte-associated 1 homolog isoform X1 | 48.633211 | 0.1024060 | -0.0260643 | 0.0572123 | 1.0129798 |
| GB50303 | unc-112-related protein-like isoform 1 | 48.557126 | 0.0148390 | -0.0282174 | -0.0320966 | -0.0198174 |
| GB48630 | JNK1/MAPK8-associated membrane protein-like | 48.447486 | 0.1865434 | 0.1182197 | -0.0520282 | 0.0503337 |
| GB54184 | bridging integrator 3 homolog | 48.279512 | 0.2466236 | 0.0417203 | 0.0903113 | 0.0893227 |
| GB47881 | signal peptidase complex catalytic subunit SEC11A | 48.132542 | 0.5259525 | 0.0728970 | -0.0701575 | 0.6412736 |
| GB51264 | glutamine-dependent NAD(+) synthetase, transcript variant X3 | 48.094964 | 0.0142024 | -0.0534230 | -0.0001249 | -0.0485017 |
| GB41631 | 60S ribosomal protein L34 isoform X2 | 48.040862 | 0.5177244 | 0.1056566 | -0.1693279 | -0.1979203 |
| GB52789 | 60S ribosomal protein L22 isoform 1 | 48.019156 | 0.5070319 | -0.0093149 | 0.0130311 | 0.4249085 |
| GB41198 | CDGSH iron-sulfur domain-containing protein 2 homolog isoform X1 | 47.967191 | 0.3082537 | -0.0667401 | -0.0120996 | 0.0453404 |
| GB52256 | 60S ribosomal protein L5 | 47.584566 | 0.3701003 | 0.0480082 | -0.0066581 | 0.0173758 |
| GB47689 | deoxyhypusine hydroxylase-like | 47.480226 | 0.2358357 | 0.0216275 | 0.9671095 | 0.2023949 |
| GB44646 | uncharacterized protein LOC100577295 | 47.302129 | 0.3302109 | 0.1785077 | 0.0911463 | 0.0268888 |
| GB51031 | uncharacterized protein LOC727650 isoform X1 | 47.280388 | 0.5050479 | 0.1136070 | 0.0952381 | -0.0658795 |
| GB55528 | 26S proteasome non-ATPase regulatory subunit 6-like | 47.151562 | 0.3463811 | -0.0554685 | -0.2101987 | 0.0554781 |
| GB55891 | diphosphoinositol polyphosphate phosphohydrolase 1 | 47.112910 | 0.4242604 | 0.0298030 | 0.0465558 | 0.0249876 |
| GB54243 | LOW QUALITY PROTEIN: carbonyl reductase [NADPH] 1-like | 46.880407 | 0.8190460 | -0.0922616 | 0.5122605 | 0.0553386 |
| GB52946 | CCR4-NOT transcription complex subunit 7-like isoform X2 | 46.803434 | 0.4343569 | 0.1315751 | 0.0491481 | -0.0073155 |
| GB48172 | GDP-mannose 4,6 dehydratase-like isoform X3 | 46.740558 | 0.3111105 | 0.1665557 | -0.2324340 | -0.1158285 |
| GB44147 | 60S ribosomal protein L15 | 46.714214 | 0.4978002 | 0.0498709 | 0.2427027 | -0.0199072 |
| GB49750 | leucine-rich repeat neuronal protein 1-like isoform X2 | 46.626903 | 0.6966922 | 0.1934088 | -0.1826017 | -0.0551242 |
| GB40882 | 40S ribosomal protein S13 isoform X1 | 46.598165 | 0.4453960 | 0.1071488 | -0.0564354 | -0.0494756 |
| GB44803 | glutathione S-transferase omega-1 isoform X1 | 46.504648 | 0.7204112 | 0.1338771 | -0.0209039 | 0.1151655 |
| GB54165 | signal recognition particle subunit SRP68 | 46.494918 | 0.3128175 | 0.0216995 | -0.1242084 | -0.2011966 |
| GB49364 | splicing factor U2af 38 kDa subunit | 46.472765 | 0.5054174 | 0.0178074 | 0.6264466 | 0.1099871 |
| GB51359 | 60S ribosomal protein L27a isoform X1 | 46.464438 | 0.7097672 | -0.0024837 | -0.1914487 | -0.2088384 |
| GB46774 | dnaJ protein homolog 1-like | 46.337676 | 0.1484461 | 0.1210105 | -0.0467028 | -0.1039836 |
| GB51727 | activator of 90 kDa heat shock protein ATPase homolog 1-like isoform 2 | 46.273632 | 0.4366550 | 0.0956855 | -0.0363729 | 0.0126642 |
| GB48886 | protein YIF1B-like | 46.028119 | 0.5613595 | -0.1968543 | 0.6536449 | 0.0233382 |
| GB47030 | serine/threonine-protein phosphatase 6 catalytic subunit isoform 1 | 45.990010 | 0.4341155 | 0.0759850 | -0.0504535 | 0.1418442 |
| GB44927 | DNA excision repair protein haywire isoform X2 | 45.979563 | 0.1517190 | 0.0417968 | 0.0208726 | 0.1384257 |
| GB48261 | 116 kDa U5 small nuclear ribonucleoprotein component-like isoform 1 | 45.925218 | 0.2817736 | 0.1651438 | 0.7974310 | -0.0546132 |
| GB53247 | transmembrane emp24 domain-containing protein-like | 45.867920 | 0.5408780 | -0.0406452 | -0.0698274 | -0.0735425 |
| GB41362 | H/ACA ribonucleoprotein complex subunit 1-like | 45.853707 | 0.4084010 | 0.1768990 | -0.1266717 | -0.0936333 |
| GB40395 | transmembrane emp24 domain-containing protein bai | 45.718197 | 0.3775638 | 0.0038366 | -0.0011633 | -0.0912656 |
| GB44520 | uncharacterized protein LOC552106 isoform X1 | 45.565831 | 0.5595378 | 0.1096068 | 0.2488975 | -0.0783400 |
| GB48750 | F-box-like/WD repeat-containing protein ebi isoform 1 | 45.555075 | 0.5668867 | 0.1738535 | -0.0462195 | 0.1490442 |
| GB50513 | dihydropteridine reductase isoform X1 | 45.519610 | 0.4696026 | -0.0805356 | -0.1389577 | -0.0478528 |
| GB44905 | serine/threonine-protein kinase mTOR | 45.440451 | 0.1399487 | -0.0801889 | 0.0358749 | -0.0938493 |
| GB54343 | 10 kDa heat shock protein, mitochondrial-like isoform X1 | 45.414153 | 1.3670969 | 0.3058799 | -0.1385513 | -0.1028236 |
| GB54131 | uncharacterized protein LOC726184 | 45.096310 | 0.2581199 | -0.0324977 | 0.0064764 | -0.1899681 |
| GB55288 | O-acetyl-ADP-ribose deacetylase MACROD2-like isoform X3 | 45.010043 | -0.0701100 | 0.0501111 | -0.0148078 | 0.0669644 |
| GB40513 | GPI transamidase component PIG-S-like isoform X1 | 44.900368 | 0.2520018 | -0.0029193 | 0.0565069 | -0.0221804 |
| GB41525 | 3-ketodihydrosphingosine reductase-like isoform 1 | 44.781531 | 0.3324147 | -0.1206715 | -0.0847338 | -0.0189033 |
| GB46375 | ubiquitin-conjugating enzyme E2 variant 2-like isoform 1 | 44.762194 | 0.5367427 | 0.1156024 | 0.0835825 | 0.0435480 |
| GB45624 | diphthine synthase | 44.598973 | 0.6656462 | 0.1686916 | -0.1192467 | 0.1462904 |
| GB43379 | membrane-bound transcription factor site-2 protease-like | 44.339755 | 0.6085154 | -0.0932733 | -0.0354369 | 0.1984883 |
| GB47590 | 40S ribosomal protein S7 | 44.263419 | 0.5270456 | 0.0158844 | -0.0091037 | 0.0814972 |
| GB44841 | methylthioribose-1-phosphate isomerase-like isoform X4 | 44.167984 | 0.6414365 | 0.2232774 | -0.1929597 | 0.0307568 |
| GB46462 | 6-phosphogluconolactonase-like | 44.116737 | 0.4650669 | 0.0373244 | -0.0835065 | -0.8228923 |
| GB44870 | zinc finger protein 706-like isoform X3 | 43.891333 | 0.3545765 | 0.1076419 | 0.0446852 | 0.0675200 |
| GB43115 | U6 snRNA-associated Sm-like protein LSm7 isoform X3 | 43.467445 | 0.4776964 | -0.0135304 | -0.0478167 | -0.1758477 |
| GB43999 | peroxiredoxin-5, mitochondrial | 43.349254 | 0.0732070 | -0.3380318 | -0.3673372 | -0.2759739 |
| GB42809 | translationally-controlled tumor protein homolog isoform 1 | 43.010403 | 0.4061440 | 0.0698239 | 0.0900855 | 0.0633612 |
| GB45374 | 40S ribosomal protein S23-like | 42.727967 | 0.5758931 | 0.1610627 | -0.0270890 | 0.2803566 |
| GB49552 | venom protease | 42.578657 | 0.1675165 | 1.4648481 | -0.0215227 | -0.0318005 |
| GB50228 | uncharacterized protein LOC726353 isoform X1 | 42.577074 | 0.1211343 | -0.1284816 | 0.3793189 | -0.0821865 |
| GB44678 | hairless | 42.238089 | 0.2916253 | 0.1073772 | 0.0558900 | -0.0093131 |
| GB44936 | histone-arginine methyltransferase CARMER-like isoform 1 | 42.203187 | 0.3900523 | -0.0439618 | 0.0049739 | 0.2849423 |
| GB53070 | phosphatidylcholine:ceramide cholinephosphotransferase 1-like isoform X1 | 42.186019 | 0.2529439 | 0.0004315 | 0.0472357 | 0.1726479 |
| GB42705 | protein archease-like | 42.063547 | 1.1836621 | 0.0316737 | -0.0220735 | 0.0778518 |
| GB50929 | mitochondrial import receptor subunit TOM40 homolog 1-like isoform 1 | 42.005871 | 0.6013777 | 0.1121208 | -0.2549371 | -0.0340123 |
| GB43634 | congested-like trachea protein-like isoform X2 | 41.916349 | 0.3995916 | 0.0722602 | -0.0719677 | 0.1157975 |
| GB45690 | phosphoribosylformylglycinamidine synthase | 41.774341 | 0.3164739 | -0.0276276 | -0.0571260 | 0.0472686 |
| GB50873 | 60S ribosomal protein L30 isoform 1 | 41.746568 | 0.5402477 | 0.1317952 | 0.1376664 | 0.5569548 |
| GB42516 | Rab escort protein | 41.739346 | 0.2774325 | 0.2220908 | 0.0370276 | 0.0213474 |
| GB42189 | 15 kDa selenoprotein-like isoform X1 | 41.640051 | 0.3217956 | 0.0086248 | -0.0226840 | 0.1198621 |
| GB48215 | endoplasmic reticulum resident protein 44 isoform X2 | 41.597766 | 0.2290880 | 0.0486694 | -0.1194541 | 0.1039772 |
| GB55220 | L-xylulose reductase | 41.366242 | 0.4809481 | 0.1879742 | -0.0277015 | 0.0302539 |
| GB40073 | COP9 signalosome complex subunit 8-like | 41.235040 | 0.3732147 | 0.0471352 | 0.0949733 | -0.1663873 |
| GB55989 | AN1-type zinc finger protein 2B-like isoform X1 | 41.195155 | 0.2808078 | -0.1577414 | -0.3145776 | 0.1737176 |
| GB42780 | CCHC-type zinc finger protein CG3800-like isoform X3 | 40.989593 | 0.4092362 | 0.0394895 | -0.0121921 | 0.5246606 |
| GB44206 | death-associated protein 1-like | 40.987986 | 0.5192956 | -0.1178852 | -0.1665770 | 0.1888227 |
| GB43844 | ARL14 effector protein-like | 40.832929 | 0.5968430 | 0.0038058 | 0.2182950 | 0.0570652 |
| GB44735 | 2-hydroxyacylsphingosine 1-beta-galactosyltransferase-like isoform X2 | 40.832642 | 0.2204333 | 0.0730009 | -0.0484108 | 0.1718789 |
| GB49312 | glyoxylate reductase/hydroxypyruvate reductase-like | 40.826942 | 0.3504554 | -0.0231545 | -0.0319473 | -0.0845797 |
| GB49628 | cytosolic non-specific dipeptidase-like isoform 1 | 40.777943 | 0.5284877 | 0.0642127 | -0.1101325 | -0.0712679 |
| GB40866 | heat shock protein cognate 4 | 40.767345 | 0.4959365 | 0.0757019 | -0.3686440 | -0.4316138 |
| GB42829 | juvenile hormone epoxide hydrolase 1 | 40.756928 | 0.4913756 | -0.0032371 | -0.0424883 | -0.1199800 |
| GB55186 | B(0,+)-type amino acid transporter 1-like | 40.582654 | 0.4529002 | 0.0743865 | 0.0275208 | 0.0831733 |
| GB53027 | mediator of RNA polymerase II transcription subunit 4 | 40.496747 | 0.8484267 | 0.0886531 | 0.0887659 | 0.0321004 |
| GB46334 | uncharacterized protein LOC724516 | 40.373143 | 0.5353789 | 0.0786186 | 0.0594108 | -0.0320568 |
| GB51188 | lysophospholipid acyltransferase 2-like | 40.363198 | 0.8433372 | -0.1379839 | -0.1829511 | -0.0977305 |
| GB47514 | dynactin subunit 6 | 40.146181 | 0.5054052 | -0.1031793 | 0.0604960 | 0.0139110 |
| GB52102 | 40S ribosomal protein S26 | 40.131992 | 0.3976449 | 0.0070740 | 0.1111791 | -0.0226270 |
| 726117 | triosephosphate isomerase | 40.090804 | 0.1395483 | -0.1446589 | 0.1406493 | -0.1781347 |
| GB47553 | electron transfer flavoprotein subunit alpha, mitochondrial-like isoform 1 | 39.917364 | 0.6502117 | 0.0287081 | -0.0954829 | 0.4726565 |
| GB41522 | methyl-CpG-binding domain protein 2 isoformX3 | 39.872734 | 0.5590640 | 0.1441634 | -0.1323572 | -0.0203530 |
| GB54263 | integrator complex subunit 6-like isoform X2 | 39.853968 | 0.1091482 | 0.2221864 | -0.0539982 | -0.0538853 |
| GB54166 | uridine 5’-monophosphate synthase isoform X1 | 39.789943 | 0.5356910 | 0.0077522 | -0.1132544 | -0.0118682 |
| GB51062 | exostosin-1 isoform X2 | 39.767134 | 0.6700835 | -0.1649515 | -0.0674832 | -0.0065497 |
| GB50981 | enoyl-CoA delta isomerase 2, mitochondrial-like | 39.664539 | 0.4853369 | 0.0142770 | -0.1447816 | 0.1043021 |
| GB53138 | inorganic pyrophosphatase-like | 39.494639 | 0.4374424 | 0.0801488 | 0.6293370 | 0.0720627 |
| GB51533 | pleckstrin homology domain-containing family F member 2-like isoform 2 | 39.482219 | 0.3383239 | 0.0040882 | 0.0148291 | -0.0667705 |
| GB55053 | soluble calcium-activated nucleotidase 1-like | 39.368821 | 0.6681203 | -0.1064675 | -0.0743620 | 0.0515004 |
| GB55081 | TM2 domain-containing protein CG11103-like | 39.261033 | 0.2691649 | 0.1561582 | -0.0534835 | 0.0031019 |
| GB54693 | fumarylacetoacetate hydrolase domain-containing protein 2A-like isoformX2 | 39.260362 | 0.4670920 | -0.0336729 | -0.0612872 | -0.1413475 |
| GB52500 | rRNA 2’-O-methyltransferase fibrillarin | 39.249959 | 0.7049029 | 0.3018185 | 0.0747833 | -0.0542918 |
| GB47469 | rRNA-processing protein FCF1 homolog | 39.226316 | 0.3784451 | 0.0773401 | -0.1736200 | 0.0064344 |
| GB40887 | V-type proton ATPase subunit E isoform 3 | 39.169647 | 0.2660676 | -0.0541195 | -0.2188922 | 0.0628958 |
| GB50637 | SUZ domain-containing protein 1-like isoform X2 | 39.158264 | 0.3365759 | 0.1525198 | -0.0322719 | 0.1197489 |
| 413799 | mitochondrial import inner membrane translocase subunit Tim17-A isoform 2 | 39.088007 | 0.6277067 | -0.0118524 | 0.0953315 | 0.0244473 |
| GB40208 | WD40 repeat-containing protein SMU1-like isoform 1 | 38.990223 | 0.3488774 | -0.0759484 | -0.0018886 | 0.0822596 |
| GB41342 | DNA replication licensing factor Mcm7 | 38.907369 | 0.5220052 | 0.0661896 | -0.2051643 | 0.1026983 |
| GB54192 | 60S ribosomal protein L13 isoform 1 | 38.869924 | 0.3960935 | 0.0581891 | 0.0284640 | 0.1850297 |
| GB49159 | probable nuclear transport factor 2-like isoform 3 | 38.577587 | 0.7173716 | 0.1870569 | -0.0748590 | 0.0314335 |
| GB47441 | V-type proton ATPase 21 kDa proteolipid subunit-like | 38.387577 | 0.5488847 | 0.0296993 | -0.0164798 | 0.0005787 |
| GB47399 | muscle segmentation homeobox-like isoform X1 | 38.281333 | 0.7726426 | -0.2560368 | -0.4934270 | -0.1176520 |
| GB52698 | synaptobrevin-like isoformX1 | 38.199554 | 0.6054745 | 0.0700210 | -0.0062005 | 0.7656234 |
| GB41598 | brahma associated protein 55kd | 38.136391 | 0.5223367 | 0.1824931 | -0.0865195 | 0.0770117 |
| GB51590 | protein MEF2BNB homolog | 37.970941 | 0.0350231 | 0.5733454 | -0.1097075 | -0.0192604 |
| GB50299 | tubulin–tyrosine ligase-like protein 12-like | 37.863749 | 0.4386535 | 0.0200704 | 0.0203359 | 0.0931801 |
| GB42649 | putative deoxyribonuclease TATDN1-like isoform X1 | 37.767873 | 0.6144898 | -0.1334785 | -0.0008459 | -0.0464900 |
| GB55013 | NTF2-related export protein isoform X5 | 37.737791 | 0.2957515 | 0.0715186 | -0.1244231 | 0.2906846 |
| GB52120 | peroxiredoxin-6 | 37.723802 | 0.3815528 | -0.0549447 | -0.0153712 | 0.1088140 |
| 412837 | ribonuclease H2 subunit A-like | 37.637595 | 0.6143030 | 0.2112714 | 0.0226790 | -0.0337567 |
| GB48905 | glutathione S-transferase S1 | 37.552635 | 0.6149408 | 0.0735945 | 0.0441112 | 0.1725105 |
| GB45435 | LOW QUALITY PROTEIN: probable tRNA(His) guanylyltransferase-like | 37.547394 | 0.5282156 | 0.0298744 | 0.0549206 | -0.0936598 |
| GB43228 | uncharacterized protein LOC408327 isoform X1 | 37.497565 | 0.3574256 | 0.0669808 | -0.1890138 | 0.0886291 |
| GB53737 | zinc finger protein Xfin-like | 37.344477 | 0.6339409 | -0.0096106 | -0.0772934 | 0.1107826 |
| GB51043 | arginase-1-like isoform X2 | 37.281674 | 0.1954107 | -0.0820244 | -0.0722952 | -0.2933409 |
| GB51543 | 60S ribosomal protein L13a isoform 2 | 37.211310 | 0.5936986 | 0.0355861 | 0.3556922 | -0.1331714 |
| GB45012 | adenosylhomocysteinase-like | 37.172393 | 0.5826456 | 0.1557345 | -0.0089272 | 0.2478642 |
| GB43697 | mediator of RNA polymerase II transcription subunit 16 isoform X3 | 37.166590 | 0.4572046 | 0.0564145 | 0.9954842 | -0.0983094 |
| GB47617 | peptidyl-prolyl cis-trans isomerase-like | 37.129701 | 0.5692158 | 0.0566404 | 0.0179629 | 0.5436083 |
| GB45147 | clavesin-2-like | 36.835485 | 1.2746476 | -0.3386410 | 0.0210604 | -0.1062741 |
| GB42239 | senecionine N-oxygenase-like isoform X3 | 36.751052 | 0.9159164 | 0.2969766 | -0.2133698 | -0.0683422 |
| GB45640 | adenosine monophosphate-protein transferase FICD homolog isoform 1 | 36.667935 | 0.5072528 | 0.1286747 | -0.2046637 | 0.1015171 |
| GB48811 | ATP-dependent RNA helicase bel | 36.570261 | 0.3780202 | 0.0460197 | -0.1383828 | 0.0565625 |
| GB49264 | protein phosphatase methylesterase 1-like isoform 1 | 36.437393 | 0.2721547 | -0.0278315 | 0.1217598 | 0.0078390 |
| GB50928 | 39S ribosomal protein L14, mitochondrial | 36.372804 | 0.5544350 | -0.0195158 | 0.0016640 | 0.1106904 |
| GB45433 | small ribonucleoprotein particle protein B | 36.334570 | 0.6382426 | 0.2386140 | 0.0583849 | -0.1183339 |
| GB42039 | ribosomal RNA processing protein 36 homolog | 36.173318 | 0.1783264 | -0.0143247 | 1.3149644 | 0.1733263 |
| GB55282 | von Willebrand factor A domain-containing protein 9-like | 36.034725 | 0.1389616 | 0.0665630 | -0.0764848 | 0.1580834 |
| GB50304 | zinc transporter 7-like | 35.996177 | 0.6582333 | 0.0821459 | -0.1040851 | -0.0658023 |
| GB49129 | uncharacterized protein LOC408761 isoform 2 | 35.770421 | 0.2113484 | 0.1095795 | -0.5414360 | -0.0139188 |
| GB47973 | integrin-linked protein kinase-like | 35.678532 | 0.3762086 | 0.0006932 | 0.0191762 | -0.0349425 |
| GB52033 | uncharacterized protein LOC100578121 isoform X2 | 35.582917 | 0.3537501 | -0.4012048 | 0.3829831 | -0.1716993 |
| GB46059 | THO complex subunit 3-like | 35.530423 | 0.3194108 | 0.0717260 | -0.0117113 | -0.0779956 |
| GB53712 | putative phospholipase B-like lamina ancestor-like isoform X1 | 35.414923 | 0.6169889 | -0.1238982 | -0.0328792 | -0.1221804 |
| GB50198 | serine/threonine-protein kinase grp isoform X3 | 35.255701 | 0.4279161 | 0.1002945 | -0.1677843 | -0.0779089 |
| 726972 | uncharacterized protein LOC726972 isoform X2 | 35.210866 | 0.6476377 | -0.1440157 | -0.0907672 | -0.0189809 |
| GB44292 | cyclin-dependent kinase 14-like isoform X3 | 35.055190 | 0.2714366 | 0.0709247 | -0.0685493 | -0.0231703 |
| GB48364 | inactive hydroxysteroid dehydrogenase-like protein 1-like isoform X3 | 35.036875 | 0.6741367 | -0.0122741 | -0.0418749 | -0.0280792 |
| GB53628 | serine/threonine-protein kinase STK11 isoform X1 | 34.891795 | 1.2258648 | 0.1815268 | 0.2809482 | 0.0470098 |
| GB44130 | nucleolar protein 9-like | 34.810127 | 0.0665729 | -0.0372271 | -0.0167726 | 0.0511577 |
| GB52433 | BET1 homolog isoform X2 | 34.796416 | 0.5047887 | -0.1044451 | 0.0848754 | 0.1821689 |
| GB45490 | regulator of chromosome condensation 1, transcript variant X9 | 34.691978 | 0.2896307 | 0.2398020 | 0.0958906 | 0.0191185 |
| 724802 | protein Asterix-like | 34.625251 | 0.9962429 | -0.0774107 | -0.0921391 | 0.0100145 |
| GB51698 | hexamerin 70a precursor | 34.611360 | 4.0709651 | 0.1794889 | -0.2182265 | -0.2564748 |
| GB45527 | TM2 domain-containing protein almondex | 34.595298 | 0.4263687 | 0.0437405 | 0.1267896 | 0.0272550 |
| GB55515 | inositol oxygenase-like | 34.543629 | 0.4181886 | -0.0692648 | -0.1373579 | -0.2522306 |
| GB53823 | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial-like | 34.398700 | 0.7237633 | -0.0223450 | 0.0049224 | 0.0746281 |
| GB50265 | glutathione S-transferase D1 isoform X4 | 34.396160 | 0.5387060 | -0.0841277 | 0.6371068 | 0.0131081 |
| GB51977 | PEST proteolytic signal-containing nuclear protein-like | 34.369532 | 0.2503364 | 0.1323459 | 0.0669926 | 0.1923790 |
| GB43709 | cytochrome P450 9e2-like isoform X3 | 34.366441 | 0.0310104 | -0.0817461 | 0.0398331 | -0.1268613 |
| 412189 | phosphopantothenoylcysteine decarboxylase-like isoform X2 | 34.310445 | 0.2180685 | -0.0241535 | 0.0498233 | -0.1011485 |
| GB44133 | tubulin beta-1 chain | 34.216225 | 0.1425112 | 0.1583071 | -0.1100392 | -0.1070295 |
| GB40596 | ubiquitin-like protein 7-like isoform X2 | 34.191567 | 0.4242164 | 0.0705997 | -0.1479794 | -0.0407598 |
| GB46844 | uracil phosphoribosyltransferase homolog isoformX2 | 34.091116 | 0.5031466 | 0.1660574 | -0.0300010 | 0.0660326 |
| GB50902 | glyceraldehyde-3-phosphate dehydrogenase 2 isoform 1 | 34.080725 | 0.1704842 | -0.0559713 | 0.0043715 | -0.0009891 |
| GB53341 | sortilin-related receptor | 34.025613 | 0.1655368 | 0.0665988 | -0.0906773 | -0.1495009 |
| GB41142 | probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase-like isoform X2 | 34.022238 | 0.7118568 | 0.0037875 | 0.0464534 | 0.1383188 |
| GB43471 | alpha/beta hydrolase domain-containing protein 17B-like isoformX1 | 34.018729 | 0.2980576 | 0.0936578 | -0.1490984 | 0.1285026 |
| GB51283 | retinal dehydrogenase 1-like isoformX1 | 34.005792 | 0.4277303 | 0.2285386 | 0.0656507 | -0.0458237 |
| GB40976 | heat shock protein 90 | 33.999770 | 0.2842467 | 0.1271968 | -0.5039286 | 0.0374273 |
| GB52648 | Golgi SNAP receptor complex member 2 | 33.918858 | 0.2966624 | 0.1435515 | -0.1180942 | 0.0429528 |
| GB52893 | protein sel-1 homolog 1-like | 33.912818 | 0.1162447 | 0.0814928 | -0.2028604 | -0.0536227 |
| GB53540 | AP-2 complex subunit alpha isoformX1 | 33.836994 | 1.2207820 | 0.0668260 | -0.0311315 | 0.0286913 |
| GB44804 | uncharacterized protein LOC100578631 isoform X1 | 33.815745 | 0.3890033 | 0.1779732 | 0.0143412 | -0.0031628 |
| GB53656 | mitochondrial ubiquitin ligase activator of nfkb 1-like | 33.731016 | 0.3618164 | 0.1463395 | 0.3565455 | 0.2768054 |
| GB42244 | uncharacterized protein LOC100576169 | 33.593810 | 0.2262147 | -0.1343608 | -0.0763432 | -0.1179807 |
| 102654594 | WD repeat-containing protein 18-like | 33.592994 | 0.8522417 | -0.0160588 | -0.2746388 | -0.0658119 |
| GB48111 | proteasome subunit beta type-1 | 33.588540 | 0.5521815 | -0.0956113 | -0.0390874 | 0.3819071 |
| GB42747 | la-related protein 4-like isoform X2 | 33.487155 | 0.2965637 | -0.0720432 | -0.3111985 | -0.0642602 |
| GB53500 | transcriptional regulator Myc-B-like | 33.403976 | 0.5233659 | 0.2675591 | -0.2968898 | 0.0530876 |
| GB53427 | nicotinic acetylcholine receptor alpha9 subunit precursor | 33.352610 | 0.7546002 | -0.0056201 | 0.0043316 | 0.2246328 |
| GB44960 | actin-related protein 2/3 complex subunit 4 | 33.333158 | 0.5330853 | 0.0982736 | -0.0652905 | 0.0223607 |
| GB46167 | polycomb group RING finger protein 3-like | 33.308854 | 0.9574296 | 0.1014605 | 0.0599765 | 0.4142648 |
| GB51656 | zinc finger HIT domain-containing protein 3-like | 33.267316 | 0.4039773 | -0.0954720 | -0.3056184 | 0.0824175 |
| GB55139 | nucleoside diphosphate kinase | 33.259515 | 0.3924253 | -0.0414809 | -0.0310907 | -0.0381059 |
| GB44571 | PQ-loop repeat-containing protein 1-like | 33.254813 | 0.3688100 | 0.1041339 | 0.2008808 | 0.0750425 |
| GB49649 | 2-oxoisovalerate dehydrogenase subunit beta, mitochondrial-like | 33.237695 | 0.6600860 | -0.0237834 | -0.0388480 | 0.2357869 |
| GB55529 | PRA1 family protein 3-like isoform 1 | 33.229144 | 0.2353809 | 0.0090145 | 0.1193451 | 0.0700857 |
| GB41150 | 40S ribosomal protein S2 isoform 2 | 33.224993 | 0.6284082 | -0.0902920 | 0.0655730 | -0.1114302 |
| GB41465 | N-alpha-acetyltransferase 15, NatA auxiliary subunit-like isoform 1 | 32.993832 | 0.2790684 | -0.0696969 | -0.4329684 | 0.5994728 |
| 725416 | U1 small nuclear ribonucleoprotein C | 32.956622 | 0.4472242 | 0.3176340 | 0.1222915 | -0.2178911 |
| GB48225 | ras-related protein M-Ras-like isoform X1 | 32.671002 | 0.3345829 | -0.0530224 | 0.0776197 | 0.0188784 |
| GB53674 | peflin-like isoform X1 | 32.435203 | 0.3959338 | -0.0327476 | 0.1274997 | 0.1606821 |
| GB52780 | retinal rod rhodopsin-sensitive cGMP 3’,5’-cyclic phosphodiesterase subunit delta isoform X1 | 32.260049 | 0.7238365 | 0.0849793 | -0.0384044 | 0.2578750 |
| GB53440 | mitochondrial enolase superfamily member 1-like | 32.151718 | 0.1776162 | -0.1318327 | 1.5401379 | 0.1079620 |
| GB55532 | dnaJ homolog subfamily C member 2-like isoform X1 | 32.067972 | 0.2413996 | 0.1702226 | 0.0188892 | -0.0407716 |
| GB45871 | protein halfway-like isoform X2 | 32.064078 | 0.3415784 | -0.0335953 | -0.8563808 | 0.0089181 |
| GB42465 | DTW domain-containing protein 2-like isoform 1 | 31.931286 | 0.6219920 | 0.2379340 | -0.2470032 | 0.1068856 |
| GB43302 | SPARC isoformX2 | 31.878990 | 0.2619840 | 0.0263749 | 0.1496186 | 0.3523250 |
| GB51257 | eukaryotic translation initiation factor 5B | 31.875261 | 0.1044346 | 0.0730455 | -0.0365932 | -0.0068101 |
| 102656905 | nuclear receptor-binding factor 2-like | 31.761967 | 0.1086756 | -0.0803804 | 0.1474292 | 0.0384732 |
| GB54865 | TATA-box-binding protein-like | 31.749942 | 0.8660753 | -0.0173659 | -0.0978571 | -0.0816912 |
| GB42252 | armadillo repeat-containing protein 6 homolog isoform X1 | 31.702890 | 1.2709305 | 0.1845252 | -0.1368748 | -0.0675243 |
| GB50100 | copper homeostasis protein cutC homolog | 31.531884 | 1.0248974 | 0.0182128 | 0.0291729 | 0.2337146 |
| GB48435 | phosphotriesterase-related protein-like isoform X2 | 31.411113 | 0.5460466 | 0.1454967 | 0.0745007 | 0.2220091 |
| GB52251 | multifunctional protein ADE2, transcript variant X2 | 31.396200 | 1.0875039 | 0.0247651 | 0.0162857 | -0.1691127 |
| GB46916 | cyclin-related protein FAM58A-like isoform 1 | 31.382010 | 0.4234627 | 0.1109239 | -0.1763806 | 0.1182154 |
| GB41827 | ATP-binding cassette sub-family G member 5-like isoform X1 | 31.285479 | 0.4530376 | -0.1428870 | -0.0654960 | 0.0102242 |
| GB40362 | flap endonuclease 1 | 31.277116 | 0.8298649 | 0.0452845 | -0.1322030 | 0.0309338 |
| 102656183 | histidine triad nucleotide-binding protein 1-like | 31.237390 | 0.5690462 | 0.0385712 | 0.2355515 | -0.0547463 |
| GB48638 | autophagy-specific gene 6 isoform X1 | 31.232070 | 0.5681974 | -0.0265661 | 0.0543033 | 0.0584607 |
| GB40259 | sel1 repeat-containing protein 1 homolog isoform 2 | 31.212354 | 0.1376695 | -0.2944141 | 0.0555338 | 0.0710728 |
| GB50603 | ribose-5-phosphate isomerase | 31.170045 | 0.8285446 | -0.1338319 | 0.0237765 | 0.0421493 |
| GB54634 | uncharacterized protein LOC725260 isoform X1 | 31.137384 | 0.8775452 | -0.0659674 | -0.3703763 | -0.0965796 |
| GB41841 | glucoside xylosyltransferase 1-like | 30.810540 | 0.1769068 | 0.2874888 | -0.1092433 | -0.3125580 |
| GB44918 | transcription initiation factor TFIID subunit 5 isoform X1 | 30.759823 | 0.4251998 | 0.1419422 | 0.0382117 | 0.0645328 |
| GB44731 | tafazzin homolog isoformX2 | 30.738304 | 0.4114771 | 0.0114507 | -0.1192272 | -0.2164392 |
| GB41806 | calcyphosin-like protein-like isoform X1 | 30.706926 | 0.5851605 | -0.0656464 | 0.0784322 | -0.2542647 |
| GB51079 | probable UDP-glucose 4-epimerase-like | 30.649903 | 0.5580345 | 0.3067217 | 0.1699667 | 0.0331168 |
| GB46254 | NAD-dependent protein deacetylase Sirt4 isoform 2 | 30.410241 | 0.1273520 | -0.1698665 | 0.1228588 | 0.1010001 |
| GB53419 | nicotinamidase-like isoform X3 | 30.372005 | 0.0893133 | -0.1542998 | -0.1407974 | -0.2210602 |
| GB50527 | putative aminopeptidase W07G4.4-like | 30.180623 | 0.4359569 | -0.0364170 | 0.0989020 | -0.1947446 |
| GB46544 | DNA-directed RNA polymerase II subunit RPB7-like | 30.054552 | 0.1100188 | 0.1691845 | 0.0778603 | 0.1406468 |
| GB49521 | NAD(P)H-hydrate epimerase-like isoform 1 | 29.929004 | 0.5741302 | -0.0080668 | 0.0208250 | 0.1272394 |
| GB53965 | uncharacterized protein LOC100578606 isoform X5 | 29.892683 | 0.3983250 | 0.0333526 | 0.0178619 | 0.0662521 |
| GB40831 | serine protease gd isoform X2 | 29.878436 | 0.0510234 | 0.0933828 | -0.1035422 | -0.1703292 |
| GB49839 | sorting nexin-8-like isoform X2 | 29.787022 | 0.2056523 | 0.0651203 | 0.1055640 | -0.2936047 |
| GB51600 | 6-pyruvoyl tetrahydrobiopterin synthase-like | 29.668592 | 0.3046772 | 0.2902129 | -1.9326712 | 0.0575645 |
| GB46347 | uncharacterized protein LOC100577486 | 29.664994 | 0.6622822 | 0.0311075 | -0.0655065 | -0.2490207 |
| GB55816 | GTP-binding protein Rhes-like | 29.642993 | 0.4987564 | 0.2343088 | 0.0027367 | 0.2108746 |
| GB48765 | uncharacterized protein LOC410179 | 29.570101 | 0.4766588 | 0.1655350 | 0.3294756 | 0.0338604 |
| GB49808 | uncharacterized protein LOC100578801 | 29.568225 | 0.7269238 | 0.2352185 | -0.0187932 | 0.0719461 |
| GB54174 | E3 ubiquitin-protein ligase RING1 isoform 1 | 29.510672 | 0.6425785 | 0.0440419 | 0.7884749 | -0.0686108 |
| GB41901 | protein phosphatase PTC7 homolog | 29.428359 | 0.2094565 | 0.0834667 | 0.0939843 | 0.0300697 |
| GB51964 | F-box/LRR-repeat protein 4 isoform X1 | 29.414811 | 0.1934627 | 0.1246155 | 0.1516790 | 0.1638128 |
| GB51088 | isovaleryl-CoA dehydrogenase, mitochondrial-like | 29.407560 | 0.2242543 | 0.1480175 | -0.0697042 | -0.1237095 |
| GB48566 | BTB/POZ domain-containing protein 2-like isoform X2 | 29.307070 | 0.6609997 | 0.0140202 | -0.0985550 | 0.0814511 |
| GB53925 | uncharacterized protein LOC724993 | 29.103857 | 0.5088155 | -0.0219957 | 0.0137111 | -0.0592972 |
| GB48983 | RING finger protein 121-like isoform X3 | 28.981301 | 0.5407186 | 0.0156023 | -0.1881486 | 0.0552048 |
| GB50427 | nucleoporin NDC1-like | 28.973708 | 0.6836237 | 0.3949079 | -0.0816620 | -0.1138518 |
| GB50087 | solute carrier family 25 member 44-like isoform X1 | 28.888649 | 0.1056903 | -0.1144416 | -0.0343900 | -0.0622543 |
| GB48250 | putative gamma-glutamylcyclotransferase CG2811-like isoform X4 | 28.883100 | 0.4355265 | -0.0197162 | -0.0314356 | 0.1135341 |
| GB46249 | non-structural maintenance of chromosomes element 1 homolog isoform X1 | 28.851656 | 0.2092181 | -0.0888038 | 0.0184521 | 0.1300475 |
| GB55496 | pyruvate dehydrogenase E1 component subunit beta, mitochondrial | 28.836512 | 0.0111701 | -0.1652602 | -0.0961041 | 0.0087354 |
| GB54611 | antithrombin-III | 28.817759 | 0.7560407 | -0.3379042 | -0.3660910 | 0.7450817 |
| GB45316 | tetratricopeptide repeat protein 8-like isoformX1 | 28.810093 | 0.7312728 | -0.4271400 | 0.2064960 | 0.0312159 |
| GB55984 | protein RCC2 homolog | 28.338670 | 0.3134334 | 0.3528683 | -0.0094091 | -0.2586981 |
| GB51210 | lateral signaling target protein 2 homolog | 28.269582 | 0.0033017 | -0.0672044 | -0.0875045 | -0.0758575 |
| GB42924 | zinc transporter ZIP1-like | 28.158372 | 0.4218655 | 0.0024998 | -0.0998613 | -0.0896504 |
| GB49080 | nose resistant to fluoxetine protein 6-like isoform X1 | 27.955088 | 0.7965459 | -0.2482813 | 0.6468446 | -0.2593906 |
| GB45850 | clavesin-2 isoform X1 | 27.917818 | 0.3462547 | 0.1786017 | -0.0403362 | 0.0571887 |
| GB40906 | myb-like protein P-like isoform X2 | 27.822077 | 0.2541050 | -0.3743120 | -0.2370745 | -0.0554477 |
| GB54485 | UDP-glucuronosyltransferase 1-3-like | 27.810168 | 0.2278894 | 0.4785468 | 0.1038331 | 0.0755244 |
| GB43256 | ATP-binding cassette sub-family D member 1-like | 27.776357 | 0.7924238 | 0.0110735 | 0.0954340 | -0.1462086 |
| GB44109 | peptidylglycine alpha-hydroxylating monooxygenase | 27.772170 | 0.2974999 | 0.1141963 | 0.0420266 | -0.0499873 |
| GB49013 | RNA-binding protein 8A | 27.750317 | 0.7522450 | -0.0250096 | 1.3311638 | 0.0345261 |
| GB47955 | S-adenosylmethionine synthase-like isoform X1 | 27.739687 | 0.0151463 | 0.0697443 | 0.0491890 | -0.8258631 |
| GB44798 | uncharacterized protein LOC410725 isoform X2 | 27.686933 | 0.1437405 | -0.0707626 | 0.4714857 | -0.0286064 |
| GB46017 | uncharacterized protein C4orf29 homolog isoform X3 | 27.642151 | 0.5102835 | 0.1383028 | 0.0603317 | 0.0278043 |
| 726289 | transcription factor AP-1 | 27.304862 | 0.4485677 | -0.0314476 | 0.1690213 | -0.0536848 |
| GB49331 | leucine-rich repeat neuronal protein 1-like | 27.177192 | 0.7165005 | 0.1190606 | 0.1251638 | -0.1974832 |
| GB44871 | glycine N-methyltransferase-like | 27.121342 | 0.1565539 | -0.1719917 | 0.3473429 | -0.3408810 |
| GB43816 | probable tubulin polyglutamylase TTLL1-like isoform X1 | 27.090329 | 0.6247142 | -0.2125553 | 0.2205965 | 0.1756876 |
| 726860 | cytochrome b5-like isoform 1 | 27.054590 | 0.7781139 | -0.0943622 | 0.0442322 | 0.0956303 |
| GB41823 | pachytene checkpoint protein 2 homolog | 26.978707 | -0.0038771 | 0.2146061 | -0.2143731 | 0.2723933 |
| GB45700 | serine protease easter | 26.877951 | 0.1450440 | -0.2263427 | -0.0134006 | -0.0597830 |
| 411552 | ceramide glucosyltransferase | 26.819149 | 0.1868746 | -0.0843795 | -0.1422361 | -0.1427414 |
| GB54511 | probable ATP-dependent RNA helicase DDX17-like | 26.805777 | 0.1707317 | 0.0287610 | -0.4040921 | -0.0180637 |
| GB50678 | 2-oxoglutarate and iron-dependent oxygenase domain-containing protein 1-like isoform X1 | 26.786549 | 0.5544786 | 0.1773563 | 0.0155991 | -0.0061252 |
| GB46297 | cuticular protein 14 precursor | 26.754073 | 0.4781626 | -0.5100832 | 0.0415980 | -0.3862613 |
| GB54541 | leukocyte elastase inhibitor-like isoform X1 | 26.716280 | 0.3035549 | -0.4191563 | -0.1065526 | -0.0195558 |
| GB40490 | inhibitor of growth protein 5-like | 26.560746 | 0.4942504 | 0.0790253 | 0.0102877 | 0.0778037 |
| GB49403 | ragulator complex protein LAMTOR3-A-like | 26.511371 | 0.5993310 | -0.0989575 | 0.0962456 | 0.1230149 |
| GB40931 | uncharacterized protein LOC409781 isoform X2 | 26.463120 | 0.4805960 | -0.1312670 | 0.1538417 | -0.1986780 |
| GB44344 | uncharacterized protein LOC100576497 isoform X1 | 26.402541 | 0.7218161 | -0.1572443 | 0.4768652 | 0.0444223 |
| GB55973 | dentin sialophosphoprotein-like isoform X1 | 26.349769 | 0.4036062 | -0.5672132 | 0.2376085 | -0.1456705 |
| GB48335 | 40S ribosomal protein S19a-like | 26.255508 | 1.9310966 | 0.1137886 | 0.1388761 | -0.0301033 |
| GB51095 | cryptochrome 2 isoform X4 | 26.233786 | 0.0379224 | -0.0010832 | -0.3590930 | 0.1773192 |
| GB50867 | cell differentiation protein RCD1 homolog isoform X2 | 26.207232 | 0.7943851 | 0.2055238 | -0.0219147 | 0.0974795 |
| GB41224 | mediator of RNA polymerase II transcription subunit 18 isoform X1 | 26.166928 | 0.7639701 | 0.0324939 | -0.0068027 | 0.0421223 |
| GB40157 | uncharacterized protein LOC408421 isoformX2 | 26.082219 | 0.2366776 | -0.1198910 | 0.2234059 | -0.2108447 |
| GB46767 | UPF0553 protein C9orf64 homolog | 26.007129 | 0.5959738 | 0.1278340 | -0.1139575 | -0.2677754 |
| GB44559 | probable small nuclear ribonucleoprotein Sm D1-like | 25.974615 | 0.8064139 | 0.0669506 | 0.0615713 | 0.0726980 |
| GB50824 | protein trapped in endoderm-1-like isoform X3 | 25.860783 | 1.3440660 | 0.0108216 | -0.0176470 | 0.0675029 |
| GB50677 | lipoma HMGIC fusion partner-like 2 protein-like isoform X2 | 25.829936 | 0.3349662 | 0.2498731 | -0.2060702 | -0.2283655 |
| GB46215 | ras-related protein Rab-24-like isoform X2 | 25.810947 | 0.3587997 | 0.0765448 | 0.1076939 | 0.2626888 |
| GB46684 | monocarboxylate transporter 3-like | 25.620508 | 0.1898639 | -0.1036857 | -0.0372247 | 0.0665315 |
| GB48452 | protein Smaug homolog 1-like | 25.585377 | 0.0687841 | 0.1826631 | -0.0684584 | 0.1423131 |
| GB47833 | transmembrane protein 165-like | 25.501588 | 0.4242674 | 0.2512318 | 0.7126863 | 0.1263984 |
| GB41720 | uncharacterized protein LOC727121 isoform X1 | 25.224127 | 0.2649846 | 0.1637064 | 0.0271937 | -0.1419746 |
| GB48884 | egl nine homolog 1-like | 25.152847 | 0.2873459 | 0.1642512 | 0.1052568 | 0.0571584 |
| GB55629 | capa receptor-like GPCR | 25.008182 | 0.6529393 | 0.2371041 | 0.0746141 | -0.0716892 |
| GB44868 | uncharacterized protein LOC409307 | 24.919307 | 0.4300665 | 0.3894421 | -0.2051877 | 0.1249847 |
| GB48419 | carbohydrate sulfotransferase 11-like isoform X1 | 24.861867 | 0.0924938 | -0.1157845 | -0.0051810 | -0.2042507 |
| GB49026 | ataxin-2 homolog isoform X4 | 24.857003 | 0.1895306 | 0.0237818 | -0.0972415 | 0.0514067 |
| GB41222 | G-protein coupled receptor Mth2-like | 24.829642 | 0.4023854 | 0.0130028 | -0.0182808 | 0.1325339 |
| GB51611 | latrophilin Cirl-like isoform X9 | 24.676624 | 0.2225114 | -0.0837081 | 0.1235594 | -0.0707102 |
| GB50893 | insulin-like growth factor-binding protein complex acid labile subunit-like | 24.612551 | 0.0915730 | 0.1110917 | 0.0723576 | -0.2334804 |
| GB44424 | lipoma HMGIC fusion partner-like 3 protein-like isoform X2 | 24.500388 | 0.4686285 | -0.4087396 | 0.0144542 | -0.0681400 |
| GB52511 | probable methyltransferase BTM2 homolog | 24.493373 | 0.2856529 | -0.1519408 | -0.1302534 | 0.2680772 |
| GB47043 | succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial isoform X2 | 24.477285 | 0.1309517 | 0.0010817 | 1.0619318 | 0.2987727 |
| GB55096 | NADP-dependent malic enzyme isoform X3 | 24.441188 | 1.4834020 | -0.1389430 | 0.0401010 | -0.0665408 |
| GB50509 | nimrod C2 isoform X3 | 24.440592 | 0.4141087 | 0.0748913 | -0.0915857 | -0.2470926 |
| GB50218 | ornithine aminotransferase, mitochondrial | 24.437909 | 1.0004847 | 0.1929614 | 0.1480193 | 0.1135575 |
| GB47823 | CD81 antigen isoform X1 | 24.195404 | 0.1827757 | 0.1509973 | 0.2598396 | -0.1960760 |
| GB45950 | organic cation transporter protein-like isoform X5 | 24.117976 | 0.3847965 | -0.2660872 | -0.0637336 | -1.0512509 |
| GB56027 | prothrombin | 24.074828 | 0.6439574 | -0.0380335 | 0.2002270 | -0.0224381 |
| GB52258 | peptidyl-prolyl cis-trans isomerase-like 3-like | 23.961526 | 0.4829926 | -0.1145227 | 0.1310623 | 0.1013030 |
| GB41159 | bifunctional dihydrofolate reductase-thymidylate synthase | 23.959629 | 0.6645720 | 0.1681940 | 0.1631102 | -0.2037979 |
| GB41897 | GTP-binding protein 128up-like isoform 1 | 23.747737 | 0.4305934 | 0.2081510 | -0.1112843 | -0.0523071 |
| GB53381 | cytochrome c oxidase assembly protein COX11, mitochondrial-like isoformX2 | 23.638929 | 0.2774799 | -0.0811438 | 0.0101308 | 0.0719343 |
| GB41881 | U3 small nucleolar ribonucleoprotein protein IMP4-like | 23.436648 | 0.2777885 | -0.0399905 | -0.2910851 | -0.1787689 |
| GB46762 | cyclin-dependent kinases regulatory subunit-like | 23.409520 | 0.6991755 | 0.3029322 | -0.0807622 | 0.1458233 |
| GB55944 | trafficking protein particle complex subunit 6B-like | 23.290718 | 0.1946912 | 0.0800499 | 0.8888456 | 0.1964253 |
| GB43738 | phenoloxidase subunit A3 | 23.274415 | 0.8178992 | -0.0015525 | -0.1105374 | 0.1258630 |
| GB40085 | carboxypeptidase B-like | 23.162645 | -0.0880914 | 0.5498435 | -0.4701129 | -0.6711759 |
| GB10293 | aubergine | 22.990016 | 0.3603924 | 0.2336865 | 3.1250154 | 0.0223516 |
| GB50891 | solute carrier organic anion transporter family member 5A1-like | 22.950913 | 0.0205965 | -0.0346673 | 0.0484672 | -0.1399681 |
| GB53136 | chromatin assembly factor 1 subunit B | 22.892182 | 0.4492767 | 0.1760773 | -0.0081902 | 0.2573357 |
| GB45250 | uncharacterized protein LOC409595 | 22.835541 | 0.1616667 | -0.1337739 | 4.8113488 | 0.0167603 |
| 726409 | peptidyl-prolyl cis-trans isomerase H-like | 22.720396 | 0.3241290 | 0.2047631 | 0.1482712 | 0.1857630 |
| GB40778 | UDP-galactose translocator | 22.704589 | 0.3352234 | -0.1043535 | 0.0475606 | -0.0133141 |
| GB41376 | J domain-containing protein-like isoform 2 | 22.701178 | 0.2801721 | -0.3909910 | -0.1688615 | -0.7719860 |
| GB46832 | lactosylceramide 4-alpha-galactosyltransferase-like isoform X2 | 22.685977 | 1.2370613 | 0.2794976 | 0.0738181 | -0.1103460 |
| 726804 | protein BTG2-like | 22.534699 | 0.2013436 | 0.0036074 | 0.0249574 | 0.0546210 |
| GB44994 | coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial-like | 22.519335 | 0.2836273 | 0.0711428 | 0.1321522 | 0.0182785 |
| GB48474 | probable chitinase 3-like | 22.510089 | 0.3170042 | -1.0892794 | -0.1370133 | -0.2971068 |
| GB41912 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase-like | 22.461304 | 2.0837612 | 0.4404114 | -0.2127462 | -0.2862259 |
| GB50226 | transferrin 1 precursor | 22.312751 | 1.2337829 | -0.4342198 | -0.1939724 | -0.1412922 |
| GB41670 | sialin-like isoform X2 | 22.226273 | 0.4541402 | 0.0867647 | 0.0660579 | -0.2317839 |
| GB53750 | UPF0454 protein C12orf49 homolog isoform X2 | 22.213256 | 0.6817408 | -0.1463571 | 0.2551895 | 0.0210236 |
| GB42038 | WD repeat-containing protein 92-like | 22.040656 | 0.3330190 | -0.0430653 | -0.1517129 | 0.3275360 |
| GB42614 | LIX1-like protein-like isoformX2 | 21.905475 | 0.4260895 | 0.2255945 | 0.1146797 | 0.0634728 |
| GB48251 | [Pyruvate dehydrogenase (acetyl-transferring)] kinase, mitochondrial isoform X4 | 21.796424 | 0.2874702 | -0.0168522 | 0.0941125 | 0.0924191 |
| GB54097 | protein Malvolio isoform X3 | 21.777543 | 0.9121160 | -0.6312734 | -0.0017033 | 0.0588475 |
| GB55576 | uncharacterized LOC408661, transcript variant X2 | 21.274156 | 0.3916209 | 0.0581601 | -0.4809768 | -0.0807577 |
| GB50819 | microtubule-associated protein futsch-like isoform X2 | 21.207395 | 0.1702950 | 0.2340726 | 0.0202836 | -0.0006498 |
| GB50239 | short/branched chain specific acyl-CoA dehydrogenase, mitochondrial-like | 21.158472 | 0.7946267 | -0.0053800 | -0.2844090 | 0.0030283 |
| GB42822 | chitooligosaccharidolytic beta-N-acetylglucosaminidase-like | 20.977118 | 0.0978782 | 0.1788705 | 0.1566748 | -0.1217243 |
| GB42306 | ATP-dependent RNA helicase vasa | 20.807569 | 0.6286335 | 0.3085074 | -0.0439372 | 0.0310578 |
| GB41083 | ras-related protein Rab-23 isoformX2 | 20.362388 | 0.1417700 | 0.1167938 | 0.0131733 | 0.1229782 |
| GB55388 | arylsulfatase J-like | 20.360747 | 0.4224041 | 0.1317412 | 0.0132846 | 0.2447127 |
| GB49919 | MATH and LRR domain-containing protein PFE0570w-like isoform X2 | 20.270881 | 0.6524171 | 0.2988537 | 0.5303577 | 0.1968502 |
| GB55082 | protein PBDC1-like | 20.151629 | 0.7263361 | 0.2790082 | 0.0025923 | 0.4892421 |
| GB44079 | menin-like | 19.990028 | 0.6334125 | 0.1343951 | 0.2058177 | -0.0462450 |
| GB46686 | uncharacterized protein LOC411065 isoform X2 | 19.985390 | 0.0484926 | 0.0667052 | -0.1570095 | -0.1514708 |
| GB53956 | uncharacterized protein C17orf104-like | 19.932790 | 0.2594661 | 0.1349512 | -0.5530299 | 0.1632245 |
| GB55615 | cell wall protein IFF6-like | 19.754017 | 0.5948476 | -0.2442130 | 0.1699421 | -0.3776250 |
| GB43789 | tubulin polyglutamylase TTLL4-like isoform X6 | 19.630384 | 0.2402280 | 0.1653948 | -0.0854485 | -0.0199505 |
| GB43963 | tyrosine kinase receptor Cad96Ca-like | 19.602162 | 0.1385403 | 0.0456529 | -0.1090652 | 0.3527115 |
| GB44064 | uncharacterized protein LOC725703 | 19.595955 | 0.1753559 | -0.4976784 | 0.6518392 | -0.3025440 |
| GB50661 | inosine triphosphate pyrophosphatase-like isoform X3 | 19.564632 | 0.6086716 | 0.1821689 | 0.8561824 | -0.0819466 |
| GB46206 | LOW QUALITY PROTEIN: origin recognition complex subunit 2-like | 19.550308 | 0.0655212 | 0.0549359 | -0.1491518 | 0.3027507 |
| GB42190 | protein arginine N-methyltransferase 5 isoform X2 | 19.040369 | 0.2059919 | 0.1715484 | 0.0878941 | -0.1018650 |
| GB40837 | alpha-tocopherol transfer protein-like isoform X1 | 19.037232 | 2.0494077 | 0.2111793 | 0.3144220 | 0.0974071 |
| GB53835 | zinc finger protein 704-like isoform X4 | 19.025743 | 0.1486738 | -0.0731218 | 0.4883919 | -0.1326713 |
| GB43202 | uncharacterized protein LOC725540 isoform X2 | 18.995035 | 0.4909215 | -0.0059242 | 0.2232381 | 0.4279267 |
| GB48790 | monocarboxylate transporter 9-like isoform X4 | 18.821376 | 0.0143936 | -0.0312915 | 0.0016004 | 0.0044720 |
| GB54735 | uncharacterized protein LOC100578100 | 18.448826 | 0.6161257 | 0.0694335 | -0.3544360 | -0.4787110 |
| GB40523 | uncharacterized protein C20orf112 homolog isoform X3 | 18.401316 | 0.3561527 | -0.2782914 | 0.0116579 | -0.3352261 |
| GB55628 | probable RNA-binding protein EIF1AD-like isoform X1 | 18.256673 | 0.6025193 | 0.3214591 | 0.1051554 | 0.0819251 |
| GB55203 | yellow-e3 precursor | 18.120966 | 0.5200993 | 0.4004946 | -0.0359437 | 0.0110322 |
| GB43581 | uncharacterized protein LOC100577641 | 18.076515 | 0.4686080 | 0.0074804 | 0.1783573 | -0.1090593 |
| GB48936 | facilitated trehalose transporter Tret1-1-like | 17.945899 | -0.0549303 | 0.0206267 | 0.8995737 | -0.1952635 |
| 102654371 | pro-resilin-like | 17.867616 | 1.4636111 | 1.0042796 | 0.3007601 | 0.0144023 |
| GB42981 | beta-1,3-glucan-binding protein | 17.584820 | 0.2261198 | -0.2653720 | -0.3266416 | -0.1945631 |
| GB45015 | SHC SH2 domain-binding protein 1 homolog B-like isoform X1 | 17.427841 | 0.2039554 | 0.1067866 | -0.2983730 | 0.0477274 |
| GB48937 | facilitated trehalose transporter Tret1-like | 17.412797 | 0.3953911 | -0.0160381 | 0.0266284 | -0.3139843 |
| GB46635 | phytanoyl-CoA dioxygenase domain-containing protein 1 homolog | 17.401740 | 1.1839629 | 0.0149362 | 0.1415970 | 0.1678399 |
| GB48300 | twin protein isoform X1 | 17.308660 | 0.8254375 | 0.2486517 | -0.2057866 | 0.0298086 |
| GB42318 | uncharacterized protein LOC727116 | 16.896178 | 0.2658590 | 0.2117759 | 0.0819290 | 0.0200483 |
| GB46589 | SAGA-associated factor 29 homolog | 16.791888 | 0.2863611 | 0.1570792 | -0.4033081 | 0.0184646 |
| GB51200 | DNA-binding protein D-ETS-6-like | 16.707124 | 1.4817916 | 1.2113416 | 0.1755298 | 0.1266821 |
| GB51697 | hexamerin 70b precursor | 16.651244 | 0.9248467 | 1.5924077 | -0.1427654 | -0.3249182 |
| GB50693 | neuropeptide Y-like | 16.565203 | 0.6360874 | 0.1883506 | 0.1215497 | 0.0675225 |
| GB41669 | baculoviral IAP repeat-containing protein 5 isoform X2 | 16.330454 | 0.4048918 | 0.1856556 | -0.0364725 | -0.0010212 |
| GB51075 | putative odorant receptor 13a-like | 16.296858 | 1.9618676 | 0.6381054 | -0.5792237 | 0.2468440 |
| 551223 | probable cytochrome P450 305a1 | 16.281624 | 0.7879659 | 0.1545479 | -0.4608964 | 0.4521486 |
| GB43786 | calsyntenin-1-like | 16.095850 | 0.3820659 | -0.0888083 | 0.1999730 | -0.4354000 |
| GB41207 | 26S proteasome non-ATPase regulatory subunit 14 | 15.814350 | 0.5494013 | -0.0171297 | -0.5985089 | 0.0350963 |
| GB43778 | enhancer of split mgamma protein-like | 15.716420 | 0.4747451 | 0.1126744 | 0.1897304 | -0.1016009 |
| GB54885 | transcriptional activator cubitus interruptus isoform X1 | 15.600480 | 0.2490596 | -0.2580801 | 0.2588245 | -0.1211705 |
| GB49755 | uncharacterized protein LOC410867 | 15.313914 | 0.2579409 | 0.0158985 | 0.1922534 | 0.0041577 |
| 102654312 | KRR1 small subunit processome component homolog | 15.079442 | 0.3038765 | 0.1766635 | -0.3099838 | -0.0574668 |
| GB42865 | solute carrier organic anion transporter family member 5A1-like isoform X2 | 15.006988 | 0.3500124 | -0.2813211 | 0.0250345 | -0.1616591 |
| GB51494 | phosphoenolpyruvate carboxykinase [GTP] isoformX1 | 14.953698 | 0.4265587 | 0.0664003 | -0.1820904 | -0.0190253 |
| GB55423 | major facilitator superfamily domain-containing protein 12-like | 14.831630 | 0.0670125 | 0.1545912 | 0.1122418 | 0.3077878 |
| GB48999 | helix-loop-helix protein 11 | 14.691145 | 1.2032462 | 0.1904779 | 0.0442749 | 0.4397348 |
| GB43508 | inositol polyphosphate 1-phosphatase-like isoform X2 | 14.582376 | 0.8995039 | -0.1733001 | 0.1792418 | -0.0066540 |
| GB54549 | alpha-glucosidase precursor | 13.837729 | -0.1032902 | 0.3679270 | 0.0907531 | -0.6487863 |
| GB55482 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1-like isoform X2 | 13.271243 | 0.5441203 | 0.0848370 | -0.5433699 | -0.4133416 |
| GB51528 | DUOXA-like protein C06E1.3-like | 12.346642 | 0.3637062 | -0.1761584 | 0.2110072 | -0.3584878 |
| GB55729 | major royal jelly protein 1 | 12.059382 | 1.2663658 | 0.0540387 | 0.0529924 | 0.0860528 |
| GB41230 | TWiK family of potassium channels protein 18-like isoform X1 | 11.965445 | 0.2913849 | -0.2434501 | 0.8795289 | 0.3367857 |
| GB50469 | pituitary homeobox homolog Ptx1-like isoform X5 | 11.636791 | 0.6862815 | -0.2646145 | -0.1322774 | 0.0207802 |
| 725344 | histone H2B-like | 11.544799 | 0.8287250 | 0.2877058 | 0.0546868 | 0.3815404 |
| GB49170 | 40S ribosomal protein S15Aa-like isoform 1 | 10.727466 | 0.7575857 | 0.0485161 | 0.2191911 | -0.0479406 |
| GB42794 | circadian clock-controlled protein-like isoform 1 | 10.676774 | 0.3401701 | -0.1571344 | -0.0433361 | -0.0342076 |
| GB42580 | short-chain dehydrogenase/reductase family 9C member 7-like | 10.165651 | 0.4876963 | -0.3705932 | -0.0212293 | -0.4697422 |
| GB41967 | uncharacterized protein LOC100576746 isoform X1 | 9.971801 | 1.4321516 | 0.9766916 | 0.3786228 | NA |
| GB50477 | uncharacterized protein LOC100577527 | 9.690182 | 1.6209961 | -0.0203154 | -0.0110509 | -0.0796754 |
| GB45499 | sodium-coupled monocarboxylate transporter 2-like isoform X1 | 6.617566 | 0.3509317 | -0.3151852 | 0.4100339 | 0.4019417 |
Module 3
Table S28: List of all the genes in Module 3, ranked by their within-module connectivity, \(k\). The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
gene_list <- module_gene_list(3)
saveRDS(gene_list, file = "supplement/tab_S28.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB40980 | glucose transporter type 1 isoform X7 | 39.155203 | 0.0515493 | -0.0914463 | 1.2158850 | 0.0827688 |
| GB53562 | uncharacterized protein LOC408874 isoform X9 | 38.524205 | -0.0053583 | -0.0767846 | 0.5414695 | -0.2110189 |
| GB42840 | leukocyte receptor cluster member 8 homolog isoform X4 | 37.430874 | -0.4552272 | -0.1083006 | 0.3866756 | 0.0557920 |
| GB42142 | nuclear hormone receptor FTZ-F1 isoform X2 | 36.814881 | -0.2021608 | 0.0411590 | 0.1695697 | -0.3207699 |
| GB54291 | YTH domain-containing protein 1-like | 36.717412 | -0.2442935 | 0.0414818 | 0.3023902 | 0.0744219 |
| GB41401 | RUN and FYVE domain-containing protein 2-like isoform X4 | 36.490531 | -0.0812565 | -0.0365944 | 0.2610568 | 0.1318950 |
| GB44662 | A-kinase anchor protein 10, mitochondrial-like isoform X1 | 36.432341 | -0.2406660 | 0.0060739 | 0.2497817 | 0.0577195 |
| GB45265 | uncharacterized protein LOC409634 isoform X2 | 36.139631 | -0.4079269 | -0.0632309 | 0.2599943 | 0.1477586 |
| GB43636 | transmembrane and TPR repeat-containing protein CG4050-like isoform 1 | 35.659017 | 0.1852364 | 0.1472132 | 0.6771330 | 0.1899545 |
| GB52643 | poly(U)-specific endoribonuclease homolog | 35.336141 | 0.0061922 | 0.0672971 | 0.3553078 | -0.3260960 |
| GB46539 | protein vav-like isoform X3 | 35.216629 | -0.0659974 | -0.1911996 | 0.1640791 | -0.0902326 |
| GB53721 | receptor-type tyrosine-protein phosphatase N2-like isoform X2 | 34.716589 | -0.1851935 | 0.0959131 | 0.2832307 | 0.1999022 |
| GB52588 | conserved oligomeric Golgi complex subunit 7 | 34.709702 | -0.0513822 | -0.0465948 | -0.0373914 | 0.0821898 |
| GB50853 | myotubularin-related protein 13 isoform X4 | 33.869164 | 0.0507376 | 0.0333520 | 0.1510548 | 0.0709991 |
| GB41811 | filaggrin-like isoform X3 | 33.098429 | -0.5342210 | 0.0316090 | 0.2110495 | 0.0731692 |
| GB43218 | microtubule organizer protein 1-like isoform X4 | 33.029172 | -0.5773672 | 0.0507792 | 0.1303719 | 0.2102495 |
| GB46243 | zinc finger Ran-binding domain-containing protein 2-like isoform X2 | 32.772599 | -0.5218356 | -0.0285268 | 0.2599025 | 0.1790063 |
| GB52247 | serine/arginine repetitive matrix protein 1-like isoform X2 | 32.446726 | -0.4856359 | 0.0495576 | 0.0623353 | -0.0805552 |
| GB42651 | mitochondrial intermediate peptidase-like | 32.431912 | -0.3940520 | -0.0651001 | 0.0660068 | 0.1799557 |
| GB51503 | mitogen-activated protein kinase 1 | 32.148307 | -0.0154365 | 0.1281304 | 0.2040299 | 0.1804699 |
| GB46102 | vesicle-fusing ATPase 1-like | 31.643506 | 0.3024808 | -0.0732729 | 0.2154283 | 0.2736418 |
| GB45702 | ubiquinone biosynthesis monooxygenase COQ6-like isoform X2 | 31.608995 | -0.2582537 | -0.0284028 | 0.1919151 | 0.2482199 |
| GB45145 | MAGUK p55 subfamily member 6 isoform X4 | 31.521291 | -0.0981234 | -0.1440841 | 0.1761089 | 0.1261872 |
| GB45555 | putative RNA-binding protein 15B isoform X1 | 31.345259 | -0.4018994 | 0.0129431 | 0.1138609 | -0.0700187 |
| GB47779 | uncharacterized protein LOC410467 | 31.130864 | 0.0049755 | -0.0096486 | 0.1932615 | 0.1271564 |
| GB48526 | protein fem-1 homolog CG6966-like isoform X1 | 30.914796 | -0.0583846 | 0.1327861 | 0.0673771 | 0.1552170 |
| GB54715 | protein kintoun-like isoform X1 | 30.634038 | -0.2736121 | 0.0704394 | 0.1192902 | 0.0704214 |
| GB55514 | LOW QUALITY PROTEIN: protein suppressor of white apricot | 30.562897 | -0.3566308 | 0.1364996 | 0.0914357 | 0.0264259 |
| GB53944 | exocyst complex component 1 | 30.154087 | -0.4672611 | 0.1161820 | 0.1557686 | 0.1470324 |
| GB51596 | neuroblastoma-amplified sequence | 29.978350 | -0.6521495 | -0.0470851 | -1.0394192 | 0.2242830 |
| GB44483 | PRKC apoptosis WT1 regulator protein-like isoform X4 | 29.863103 | 0.0985655 | 0.0317316 | 0.1805171 | 0.1971958 |
| 411347 | peripheral plasma membrane protein CASK-like isoform X12 | 29.847222 | -0.2141844 | 0.1578634 | 0.2794613 | -0.3148287 |
| GB55146 | uncharacterized protein LOC409323 | 29.785985 | 0.2725906 | -0.1402887 | 0.5153834 | 0.0859016 |
| GB44208 | WD repeat-containing protein 37-like isoform X4 | 29.591743 | -0.0174274 | -0.0041616 | 0.0825576 | 0.4068904 |
| GB53700 | solute carrier family 12 member 8-like | 29.550937 | 0.0729533 | 0.0318466 | 0.1686427 | 0.0426324 |
| GB49488 | O-phosphoseryl-tRNA(Sec) selenium transferase-like isoform X3 | 28.733717 | -0.2467713 | 0.2352608 | 0.2717046 | 0.1671923 |
| GB50679 | UPF0430 protein CG31712-like isoform X3 | 28.597092 | -0.6043757 | -0.0354073 | 0.2136135 | 0.1190436 |
| GB55507 | FYVE, RhoGEF and PH domain-containing protein 4-like isoform X2 | 28.573054 | 0.1464816 | 0.1292345 | 0.1671979 | 0.0672144 |
| GB55285 | uncharacterized protein LOC724761 | 28.419308 | -0.2318332 | -0.1909534 | 0.2826152 | 0.2484804 |
| GB41146 | synembryn isoform X3 | 28.265962 | 0.0660072 | 0.0103353 | 0.1428126 | 0.1002792 |
| GB41079 | guanine nucleotide-binding protein subunit beta-5 isoform X2 | 28.079443 | -0.0150056 | 0.0661792 | 0.1762058 | 0.0499223 |
| GB49896 | kinesin 2A | 27.925674 | 0.1380803 | 0.0948071 | 0.2336285 | 0.0896094 |
| GB51002 | fasciculation and elongation protein zeta-2 isoform X4 | 27.832948 | 0.1751121 | 0.0994907 | 0.4154053 | 0.1346307 |
| GB44031 | dorsal protein isoform B | 27.826815 | -0.1264089 | -0.0969409 | 0.2227205 | -0.1212030 |
| GB44721 | cleavage and polyadenylation specificity factor subunit 5-like isoform 2 | 27.582858 | 0.0543117 | 0.0683613 | 0.0893530 | 0.2781177 |
| GB55350 | amyloid beta A4 precursor protein-binding family B member 2-like isoform X7 | 27.523852 | -0.0033611 | -0.2010763 | 0.3022455 | 0.3595586 |
| GB47963 | probable E3 ubiquitin-protein ligase HERC4-like isoform X3 | 27.279176 | -0.1072215 | 0.0085017 | 0.3024338 | 0.1582591 |
| GB44077 | kxDL motif-containing protein CG10681-like | 27.273547 | -0.0806378 | 0.1813562 | 0.1483027 | 0.1370592 |
| 102654127 | neurochondrin homolog | 27.164136 | -0.4227097 | -0.1567769 | 0.4266425 | 0.0696887 |
| GB44296 | JNK-interacting protein 1 isoform X3 | 26.897786 | -0.1116452 | -0.1539155 | 0.5432720 | 0.2334244 |
| GB50144 | galactoside 2-alpha-L-fucosyltransferase 2-like | 26.797730 | -0.6950435 | 0.0636122 | 0.4266873 | 0.2436411 |
| GB45227 | transmembrane protein 64-like isoform 1 | 26.751202 | -0.1984516 | -0.0631240 | 0.1790250 | 0.1587587 |
| GB45535 | phosphoribosyl pyrophosphate synthase-associated protein 2-like isoform X1 | 26.578051 | 0.0646934 | -0.0329508 | 0.2006897 | 0.1827454 |
| GB54201 | uncharacterized protein LOC100577050 isoform X4 | 26.192930 | -0.2625878 | 0.0378657 | 0.3161599 | 0.5298425 |
| GB49397 | insulin-like growth factor-binding protein complex acid labile subunit-like isoform X6 | 26.105822 | 0.0298056 | -0.2957700 | 0.3036323 | -0.0014611 |
| GB42675 | adenylate cyclase type 2-like | 25.971556 | -0.4302477 | -0.1156186 | 0.3942392 | 0.0897006 |
| GB52759 | brefeldin A-inhibited guanine nucleotide-exchange protein 3-like isoform X2 | 25.954786 | -0.2543826 | -0.0397308 | -0.0333180 | 0.2135026 |
| GB52464 | uncharacterized protein LOC726793 | 25.801960 | 0.2826988 | 0.0973073 | 0.0659393 | 0.2571978 |
| GB55441 | uncharacterized protein LOC409164 isoform X2 | 25.574085 | 0.1552340 | -0.0460763 | 0.1939540 | 0.0307548 |
| GB42917 | transmembrane protein 53-like | 25.514571 | -0.0089100 | 0.2011469 | 0.3120569 | 0.0961105 |
| GB45413 | GTPase activating Rap/RanGAP domain-like 3 isoform X2 | 25.513119 | -0.6745272 | 0.0814359 | 0.1845606 | 0.0041558 |
| GB55635 | cAMP-responsive element-binding protein-like 2-like | 25.489690 | -0.1137511 | -0.0119179 | 0.1326484 | 0.0432608 |
| GB46914 | unzipped precursor | 25.406465 | 0.0688776 | -0.2079623 | 0.3240089 | 0.0806107 |
| GB40973 | glutamate receptor 1-like isoform X2 | 25.188876 | 0.0343906 | -0.1784854 | 0.1815953 | 0.1776326 |
| GB52600 | uncharacterized protein LOC413002 isoform X2 | 25.018496 | -0.2291788 | 0.1380160 | 0.2663379 | 0.3479869 |
| GB45036 | breast cancer anti-estrogen resistance protein 3-like isoform X1 | 24.895367 | -0.4742001 | -0.1592545 | 0.2798366 | 0.1466932 |
| GB40305 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial isoform 1 | 24.824313 | -0.2441574 | -0.1926550 | 0.1474342 | 0.0397377 |
| GB54057 | uncharacterized protein LOC551162 isoform X3 | 24.794819 | 0.2057112 | 0.1691141 | 0.2332495 | 0.0927002 |
| GB47039 | dynamin isoform X11 | 24.654723 | -0.4514198 | -0.1258553 | 0.2012804 | 0.1285005 |
| GB55539 | uncharacterized protein LOC100577742 | 24.584418 | -0.2489971 | 0.2389540 | 0.1092647 | 0.0345600 |
| GB53582 | serine/threonine-protein kinase tricorner isoform X6 | 24.418876 | -0.3968406 | 0.0303525 | 0.0933007 | 0.1127029 |
| 408663 | soma ferritin | 24.386439 | -0.3793862 | 0.7353754 | 0.3450990 | 0.0944351 |
| GB45617 | protein eyes shut isoform X1 | 24.337514 | -0.4162517 | -0.1027993 | 0.3807624 | 0.0800984 |
| GB40343 | uncharacterized protein LOC413620 isoform X2 | 24.324698 | -0.2014450 | 0.0331841 | 0.0802108 | -0.9260997 |
| GB40356 | spondin-1-like isoform X9 | 24.093795 | 0.2385692 | 0.0173705 | 0.3801365 | 0.1357436 |
| GB45140 | junctophilin-1-like isoform X3 | 23.905069 | -0.3176739 | 0.0547538 | 0.3119804 | 0.2704978 |
| GB44523 | NACHT and WD repeat domain-containing protein 1-like isoform X2 | 23.891586 | -0.4078231 | 0.0037661 | 0.2684293 | -0.0662191 |
| GB49480 | cGMP-specific 3’,5’-cyclic phosphodiesterase-like isoform X4 | 23.856050 | 0.2547182 | -0.0856179 | -0.0945584 | 0.2762381 |
| GB48162 | anoctamin-8-like isoformX1 | 23.848473 | -0.1334123 | -0.2456800 | 0.2832485 | 0.3596429 |
| GB51722 | tetraspanin-1 isoformX1 | 23.813979 | 0.3409996 | 0.1570373 | 0.2992587 | 0.1753064 |
| GB40118 | glutamate decarboxylase-like isoform X2 | 23.789420 | -0.1639796 | -0.0828520 | 0.3842894 | 0.2501437 |
| GB40358 | RILP-like protein homolog isoform X2 | 23.607551 | -0.0595644 | 0.0531572 | 0.3154857 | 0.2155084 |
| GB51489 | proton-coupled amino acid transporter 1-like isoform X2 | 23.432554 | -0.2201609 | -0.1580994 | 0.4209838 | 0.3151434 |
| GB44746 | uncharacterized protein LOC412112 isoform X1 | 23.351191 | 0.1816134 | -0.0871766 | 0.2381982 | 0.1846010 |
| GB44482 | solute carrier family 12 member 7-like isoform X5 | 23.301647 | -0.0871783 | -0.1660832 | -1.1787562 | 0.2748877 |
| GB50415 | diacylglycerol kinase theta-like isoform X7 | 23.214175 | 0.0659766 | -0.2058608 | 0.5586630 | 0.1722711 |
| GB41746 | neogenin isoform X2 | 23.158976 | -0.3449802 | -0.1371039 | 0.1408065 | 0.0365518 |
| GB52028 | elongation factor 1-alpha | 23.107009 | -0.1320001 | 0.1115101 | 0.4240778 | 0.0393219 |
| GB43116 | sarcoplasmic calcium-binding protein 1 isoformX2 | 23.065999 | 0.4083459 | -0.0467683 | 0.1728039 | 0.3255584 |
| GB50516 | TPPP family protein CG4893-like isoform X1 | 22.733193 | 0.2592859 | -0.0017917 | 0.2166818 | 0.1251679 |
| GB43052 | paramyosin, long form-like | 22.730108 | -0.1753998 | -0.1732361 | 0.0876652 | 0.1091558 |
| GB45013 | repressor of RNA polymerase III transcription MAF1 homolog | 22.588896 | -0.0473054 | -0.0546453 | 0.2385886 | 0.1038500 |
| GB50911 | probable G-protein coupled receptor CG31760-like isoform X5 | 22.577797 | -0.6052421 | -0.3492217 | 0.2771937 | 0.2677893 |
| GB43546 | uncharacterized protein LOC410487 | 22.424375 | -0.4689972 | 0.0530247 | 0.1119586 | 0.0601744 |
| GB51838 | uncharacterized protein LOC552650 | 22.273044 | 0.0810884 | -0.4106210 | 0.1851129 | 0.2274499 |
| GB48539 | calcium-dependent protein kinase 4-like | 22.181841 | -0.3764667 | -0.0424535 | 0.1640102 | -0.6938558 |
| GB52723 | uncharacterized protein LOC726322 isoform X2 | 22.153906 | -0.3292076 | -0.0862735 | 0.3133750 | 0.2277422 |
| GB51689 | muscarinic acetylcholine receptor DM1-like | 22.101925 | 0.0815534 | 0.0500541 | -0.1721455 | 0.2141637 |
| GB40923 | nicotinic acetylcholine receptor alpha8 subunit | 22.070057 | 0.5052163 | -0.0945717 | 0.3181790 | 0.1871547 |
| GB47352 | zinc finger protein Noc-like | 21.817701 | -0.2492754 | 0.0410122 | 0.3587598 | 0.0966715 |
| GB49396 | sodium- and chloride-dependent GABA transporter 1 isoform X2 | 21.814461 | -1.3581757 | -0.1649135 | 0.8194765 | 0.0342622 |
| GB44810 | LOW QUALITY PROTEIN: carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein-like | 21.786015 | -0.3173358 | 0.0605219 | 0.3109436 | 0.1602752 |
| GB46705 | muscle M-line assembly protein unc-89 isoform X5 | 21.660134 | -0.5256014 | -0.1752851 | 0.2134925 | -0.4106971 |
| GB46673 | two pore potassium channel protein sup-9-like isoform X1 | 21.626412 | -0.1216310 | 0.1012412 | 0.1407964 | 0.1843102 |
| GB45263 | prohormone-4 | 21.606850 | 0.1538383 | -0.0323490 | 0.3624705 | 0.3772112 |
| GB48632 | synaptotagmin 14, transcript variant X3 | 21.363059 | 0.0727930 | -0.3070684 | 0.4161671 | 0.0266612 |
| GB52321 | agglutinin-like protein 1-like isoform X4 | 21.241305 | -0.3522057 | -0.0816684 | 0.3561753 | 0.2277453 |
| GB53345 | uncharacterized protein LOC100578770 | 21.092281 | -0.6699866 | 0.0478591 | 0.3629673 | 0.2243062 |
| GB40701 | uncharacterized protein LOC551765 | 21.056443 | -0.3662364 | -0.1890637 | 0.1549515 | 0.0620317 |
| GB50651 | prohormone-3 | 21.047080 | 0.1366545 | -0.0676341 | 0.3382996 | 0.2170452 |
| GB46720 | regulator of G-protein signaling 17-like isoform X1 | 20.923524 | -0.0820377 | 0.3676264 | 0.2799898 | 0.1328548 |
| GB53030 | transmembrane protein 181-like isoform X2 | 20.919242 | 0.3170529 | 0.0407814 | 0.3887276 | 0.3264278 |
| GB44369 | suppressor APC domain-containing protein 2-like isoform X2 | 20.760600 | 0.0509287 | 0.1539895 | 0.1557634 | 0.1321094 |
| GB40303 | broad-complex core protein isoforms 1/2/3/4/5-like isoform X2 | 20.629826 | 0.0741437 | -0.0687444 | 0.3963796 | 0.1786596 |
| GB49174 | uncharacterized protein LOC412265 isoform X2 | 20.586644 | -0.9324891 | 0.1064352 | 0.1906466 | -0.0504898 |
| GB52953 | guanylate cyclase, soluble, beta 1 | 20.443805 | 0.6072084 | 0.1513733 | 0.1693598 | 0.2061758 |
| GB45235 | uncharacterized protein LOC724460 | 20.276758 | -0.2838255 | 0.9094258 | 0.2583031 | 0.3126307 |
| GB40794 | synaptotagmin-4 isoform 2 | 20.255822 | -0.6093872 | -0.1360159 | 0.2684501 | 0.1741523 |
| GB44213 | filamin-like | 20.215422 | -0.4303453 | -0.0542641 | 0.4264474 | -0.0124774 |
| GB47203 | uncharacterized protein LOC552612 | 20.155226 | 0.1069055 | 0.0243062 | 0.2103766 | -0.0349762 |
| GB53340 | spectrin beta chain isoform X1 | 20.116249 | 0.2736734 | -0.0955519 | 0.3377706 | 0.0876865 |
| GB43814 | potassium channel subfamily K member 18-like isoform X2 | 20.014024 | -0.5722042 | -0.1680483 | 0.2463202 | 0.0195043 |
| 102656283 | uncharacterized protein LOC102656283 | 19.767035 | -0.5789037 | -0.0575661 | 0.8208169 | -0.0251430 |
| GB49268 | glutamate receptor ionotropic, kainate 2-like isoform X1 | 19.755842 | -0.2957918 | -0.0823437 | 0.3864055 | 0.2220661 |
| GB42529 | signal peptide peptidase-like 3-like isoform X2 | 19.664462 | 0.1323459 | -0.1493902 | 0.2219016 | 0.1617285 |
| GB48187 | mitogen-activated protein kinase kinase kinase 7 isoform X2 | 19.660413 | -0.0443926 | 0.1087015 | 0.3467619 | -0.1090669 |
| GB44988 | prohormone-2 precursor | 19.653234 | -0.0546106 | -0.2561033 | 0.3076662 | -0.7683125 |
| GB46886 | NMDA receptor 1 isoform X2 | 19.640100 | 0.3615873 | -0.2205375 | 0.3614490 | 0.2769770 |
| GB40599 | fukutin-related protein-like | 19.589442 | -0.1622265 | 0.0916666 | 0.1427301 | 0.0116238 |
| GB49732 | uncharacterized protein R02F2.2-like isoform X2 | 19.589428 | -0.5014320 | -0.1982269 | 0.1985966 | 0.0196824 |
| GB42659 | cAMP-dependent protein kinase type I regulatory subunit isoform X5 | 19.556951 | 0.0951226 | -0.1877133 | 0.6991097 | 0.0984813 |
| GB43293 | high-affinity choline transporter 1-like | 19.554643 | 0.4459734 | -0.0936936 | 0.5242129 | -0.2360701 |
| GB53652 | coenzyme Q-binding protein COQ10 homolog B, mitochondrial-like | 19.532844 | -0.3874882 | -0.2074187 | 0.0943089 | 0.0856498 |
| GB45733 | kinesin 13 isoform X2 | 19.513308 | -0.4691764 | 0.1121083 | 0.0304380 | 0.1411362 |
| GB44536 | uncharacterized protein LOC100576683 | 19.449884 | -0.5808087 | 0.0629350 | 0.2294547 | 0.1308008 |
| GB54724 | cationic amino acid transporter 4-like | 19.417278 | 0.3113255 | 0.1360071 | 0.2703558 | -0.1836218 |
| GB46165 | fibroblast growth factor receptor homolog 1 isoform X2 | 19.398835 | -0.5585660 | -0.3268543 | 0.3356287 | 0.1816028 |
| GB48892 | LOW QUALITY PROTEIN: synaptogyrin-2 | 19.361165 | -0.6324875 | 0.1682813 | 0.0625846 | 0.2719394 |
| GB54818 | muscle LIM protein Mlp84B-like isoform X2 | 19.345615 | -0.2826653 | -0.0654761 | 0.2387279 | 0.2020251 |
| GB48489 | dipeptidyl aminopeptidase-like protein 6-like isoform X1 | 19.303984 | -0.0784904 | -0.1392929 | -0.9277102 | 0.1192834 |
| GB55504 | zinc finger protein 143-like isoform X2 | 19.176118 | -0.1397815 | -0.0341459 | 0.0768685 | 0.0929361 |
| GB52763 | uncharacterized protein LOC100578776 isoform X2 | 19.167854 | -0.0468317 | -0.0133573 | 0.4850054 | 0.1898818 |
| GB41659 | endothelin-converting enzyme 1 isoform X4 | 19.100588 | -0.2768391 | 0.0360079 | 0.2676889 | 0.2457807 |
| GB55237 | disco-interacting protein 2 isoform X1 | 19.093588 | 0.1891830 | 0.0895356 | 1.4083799 | -0.8325983 |
| GB45393 | surfeit locus protein 1 | 19.046890 | -0.3007976 | -0.0407981 | 0.1662574 | 0.1525407 |
| GB47678 | protein FAM69C-like isoform X2 | 18.982397 | -0.1217844 | -0.1356090 | 0.3516168 | 0.2260748 |
| GB52679 | uncharacterized protein LOC409139 isoform X2 | 18.975709 | 0.1482778 | 0.1011442 | 0.1363535 | 0.1891389 |
| GB41866 | uncharacterized LOC552431, transcript variant X4 | 18.956451 | 0.4110198 | -0.0127336 | 0.2639560 | 0.3002171 |
| 725415 | BTB/POZ domain-containing protein KCTD16-like | 18.950874 | 0.9409643 | -0.6514749 | 0.2958511 | 0.3458427 |
| GB47990 | tropomyosin-1-like | 18.937747 | 0.1503472 | -0.3462256 | 0.1156195 | 0.3927175 |
| GB55520 | PDZ domain-containing RING finger protein 4 | 18.829258 | -0.5604969 | -0.0269463 | 0.3270404 | 0.0439836 |
| GB43818 | trimeric intracellular cation channel type B-like | 18.770242 | -0.0542100 | -0.4532958 | 0.2997434 | 0.3859680 |
| GB49969 | tubby-related protein 4-like isoform X3 | 18.752440 | -0.1399939 | 0.2723870 | 0.2337417 | 0.1057941 |
| GB43231 | uncharacterized protein LOC408329 | 18.748260 | -0.3721146 | 0.2672594 | 0.2644489 | 0.0270706 |
| GB40625 | synapsin isoform X2 | 18.745487 | -0.0007463 | 0.2642269 | 0.4957649 | 0.2952794 |
| GB42692 | ultraspiracle isoform X7 | 18.741237 | 0.0024864 | -0.0020192 | 0.2272694 | -0.2902473 |
| GB48362 | protein kinase DC2 isoform X1 | 18.697126 | 0.0144671 | -0.0862512 | 0.4479301 | 0.1956692 |
| GB54423 | uncharacterized protein LOC551958 | 18.658466 | -0.0182288 | -0.1217524 | 0.5110847 | -0.1082280 |
| GB50142 | uncharacterized protein LOC726068 | 18.625706 | -0.1832653 | 0.2964760 | 0.2214939 | 0.0568193 |
| GB55559 | D-beta-hydroxybutyrate dehydrogenase, mitochondrial-like isoform X2 | 18.615623 | 0.0097019 | -0.3884461 | 0.3465838 | 0.4558630 |
| GB44430 | dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial-like isoform X1 | 18.610733 | -0.5926677 | -0.0665588 | 0.1894260 | -0.2475919 |
| GB54446 | arginine kinase isoform X2 | 18.602183 | 0.2311390 | -0.1289730 | 0.4408448 | 0.1410691 |
| GB52013 | isocitrate dehydrogenase [NAD] subunit gamma 1, mitochondrial-like isoform 2 | 18.533786 | -0.6028590 | -0.1348051 | 0.1594659 | 0.1912483 |
| GB51787 | myosin light chain alkali-like isoform X5 | 18.437615 | 0.4439891 | -0.2793164 | 0.3723450 | 0.3497629 |
| GB47928 | allatostatins precursor | 18.420611 | -1.0599542 | -0.0880527 | 0.3043287 | 0.2816394 |
| GB40537 | reticulon-4-interacting protein 1 homolog, mitochondrial-like | 18.410422 | 0.0209046 | 0.0215747 | 0.1688833 | 0.2246331 |
| GB48163 | probable NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial isoform X1 | 18.344105 | 0.0714526 | 0.0607222 | 0.2036164 | 0.1557124 |
| GB55549 | lachesin-like isoform X1 | 18.310222 | -0.1648579 | 0.0177020 | 0.2890627 | 0.0088269 |
| GB42644 | nicotinic acetylcholine receptor alpha2 subunit precursor | 18.299345 | -0.6912923 | -0.2580954 | 0.1386431 | 0.1566976 |
| GB47663 | uncharacterized protein LOC727019 isoform X2 | 18.288701 | -0.6102756 | -0.2592534 | 0.5557404 | 0.3145607 |
| GB54598 | sodium-dependent phosphate transporter 2-like | 18.284224 | 0.4473326 | -0.0180989 | 0.2539486 | 0.0175071 |
| GB50123 | myophilin-like | 18.224951 | 0.1235467 | -0.2641748 | 0.2260472 | 0.2157248 |
| GB42676 | epimerase family protein SDR39U1-like | 18.153999 | -0.2997739 | -0.0866989 | 0.1779110 | 0.1303428 |
| GB41845 | dual specificity protein phosphatase Mpk3 | 18.146658 | -0.0481926 | 0.0168287 | 0.2460271 | 0.1511945 |
| GB41839 | uncharacterized protein LOC552552 isoform X1 | 18.134727 | -0.6290870 | -0.1294563 | 0.0826175 | -0.0749326 |
| GB51917 | 1-acyl-sn-glycerol-3-phosphate acyltransferase alpha-like isoform X1 | 18.076845 | -0.7884807 | 0.1459607 | 0.4419118 | 0.3395123 |
| GB41863 | glutamate receptor-interacting protein 1 | 17.991009 | -0.1607764 | -0.0277491 | 0.1835944 | -0.0384194 |
| GB41860 | serine/threonine-protein kinase STE20 isoform X1 | 17.951469 | -0.7087824 | 0.5159097 | 0.4174642 | 0.3735759 |
| 724169 | transcription factor kayak isoform X3 | 17.830227 | -0.3392672 | 0.0570718 | 0.3706900 | 0.3128189 |
| GB48151 | F-box/LRR-repeat protein 16 | 17.764501 | 0.2889661 | 0.0707071 | 0.4207396 | 0.2248933 |
| GB47728 | uncharacterized protein LOC409327 isoform X2 | 17.741284 | 0.0452367 | -0.0341232 | 0.5200907 | 0.2405244 |
| 410562 | serine/threonine-protein kinase SBK1-like isoform X1 | 17.708632 | -0.3271830 | 0.3895611 | 0.5692589 | -0.1388751 |
| GB48232 | uncharacterized protein LOC552578 isoform X1 | 17.668741 | -0.6357718 | -0.0956581 | 0.4937880 | 0.0942855 |
| GB45497 | histone-lysine N-methyltransferase, H3 lysine-79 specific isoform X3 | 17.637557 | -0.8636643 | -0.2042806 | 0.3040861 | -0.1837393 |
| GB40703 | cadherin-23-like | 17.589329 | 0.1546920 | -0.1493760 | -0.3526411 | 0.6811609 |
| GB44498 | uncharacterized protein LOC551934 isoform X3 | 17.498893 | 0.2086090 | -0.2604815 | 0.3855567 | 0.1219437 |
| GB41241 | SET and MYND domain-containing protein 4-like | 17.495350 | 0.0608500 | -0.1388716 | 0.3527382 | 0.0284537 |
| GB44970 | RUN domain-containing protein 1-like | 17.329197 | -0.9322910 | 0.0121563 | 0.2084180 | 0.0591890 |
| GB45406 | innexin shaking-B isoform X1 | 17.231771 | -0.5128147 | -0.1674207 | 0.5925262 | 0.1320149 |
| GB54827 | synaptotagmin 1 | 17.230302 | 0.4031080 | -0.1543626 | 0.2437063 | -0.1691167 |
| GB44889 | neurogenic protein big brain | 17.199669 | -0.6922649 | -0.0037840 | 0.3695044 | 0.5347946 |
| GB41487 | transcription factor SPT20 homolog isoform X3 | 17.170134 | -0.2291478 | -0.0286335 | 0.1732298 | 0.1687227 |
| GB40735 | fructose-bisphosphate aldolase-like isoform X1 | 17.026386 | 0.0502705 | 0.0659511 | 0.1683899 | 0.0707604 |
| GB40603 | uncharacterized protein LOC410369 | 17.025161 | -0.2494051 | -0.1161132 | 0.3930715 | -0.2010532 |
| GB50150 | major facilitator superfamily domain-containing protein 6-like isoform X2 | 17.015236 | 0.1944749 | -0.2309688 | 0.4398861 | 0.4354699 |
| GB52350 | BRO1 domain-containing protein BROX-like | 16.895098 | 0.0251370 | -0.0151419 | 0.0630647 | 0.0318475 |
| GB55976 | zinc finger protein 271-like isoform X2 | 16.891859 | 0.3351956 | 0.0037890 | 0.2225968 | 0.2054870 |
| GB49559 | intraflagellar transport protein 88 homolog isoform X2 | 16.889134 | 0.2668334 | 0.0508070 | 0.2711415 | 0.1094384 |
| GB44910 | nuclear receptor-binding protein homolog | 16.815190 | 0.0472513 | -0.0977933 | 0.1435630 | 0.1575120 |
| GB45833 | putative methyltransferase NSUN7-like isoform X1 | 16.698836 | -1.0927675 | -0.1155145 | 0.5108046 | -0.0390299 |
| GB49688 | peroxidase isoformX2 | 16.659376 | -0.1014136 | -0.2238851 | 0.1038606 | 0.0081065 |
| GB53163 | transient receptor potential channel pyrexia isoform X3 | 16.648401 | -0.3313750 | -0.3406834 | 0.2947176 | 0.2831949 |
| GB51074 | differentially expressed in FDCP 8 homolog isoform X2 | 16.564010 | -0.1425567 | 0.1420270 | 0.2989413 | 0.0375067 |
| GB48709 | uncharacterized protein LOC724300 | 16.558623 | 0.1015272 | 0.0587105 | 0.4403682 | 0.3266977 |
| GB50985 | yorkie homolog isoform X2 | 16.550915 | -0.0587983 | -0.0269504 | 0.2617774 | 0.1803131 |
| GB48331 | GTP-binding protein RAD-like isoform X2 | 16.456567 | -0.4165385 | 0.2348699 | 0.7690865 | 0.4342525 |
| GB55798 | cyclin-dependent kinase 5 activator 1 isoform X2 | 16.279053 | 0.0952047 | -0.0655147 | 0.3694129 | 0.1360139 |
| GB42775 | transmembrane protein 189-like | 16.243327 | -0.7237029 | -0.1637982 | 0.2517442 | 0.0892714 |
| GB49967 | NADP-dependent malic enzyme isoform X1 | 16.164695 | -0.4037762 | -0.0858154 | 0.2919158 | 0.2794340 |
| 412927 | uncharacterized protein LOC412927 isoform X4 | 16.110429 | -0.7754149 | -0.1198548 | 0.4751749 | 0.1089946 |
| GB47799 | protein hairy isoform X2 | 16.105701 | -0.5155486 | -0.1040845 | 0.3328577 | 0.0162919 |
| GB56017 | paired mesoderm homeobox protein 2-like isoform X2 | 16.097799 | -0.2105191 | -0.3431956 | 0.5366410 | 0.3841222 |
| GB46581 | ubiquitin-conjugating enzyme E2 H | 16.095537 | 0.0159472 | -0.0154422 | -0.1194925 | 0.0888370 |
| GB40541 | vesicular inhibitory amino acid transporter-like | 16.089482 | 0.2687177 | 0.1596356 | 0.6513254 | 0.4144100 |
| GB42792 | uncharacterized protein LOC409805 isoform X3 | 15.973436 | 0.2876909 | -0.2801362 | 0.4438630 | 0.4154002 |
| GB45017 | RNA pseudouridylate synthase domain-containing protein 2-like isoform X3 | 15.969769 | -0.3168366 | -0.0121638 | 0.2034878 | 0.2164005 |
| GB53646 | polycomb group protein Pc | 15.933332 | -0.0963384 | -0.0346554 | 0.1200365 | 0.1950194 |
| GB44011 | E3 ubiquitin-protein ligase Rnf220-like isoform X5 | 15.908808 | -0.3867762 | -0.0270429 | 0.2611436 | -0.2114363 |
| GB42757 | protein disabled isoform X4 | 15.903414 | -0.5858988 | 0.0597639 | 0.2367152 | 0.0414481 |
| GB50442 | uncharacterized protein LOC100577562 isoform X1 | 15.870343 | 0.2302594 | 0.4191878 | 0.7750161 | 0.3076271 |
| GB54037 | uncharacterized protein LOC727502 | 15.794219 | -0.1105505 | -0.1426724 | 0.8009610 | 0.0651269 |
| 410229 | slit homolog 3 protein | 15.714912 | 0.1677186 | -0.2064498 | 0.0496820 | -0.0442317 |
| GB47148 | uncharacterized protein LOC552326 | 15.713800 | 0.1051884 | 0.0235050 | 1.3461976 | 0.2395930 |
| GB53986 | dehydrogenase/reductase SDR family member 11-like isoform X1 | 15.709492 | 0.3039867 | -0.3381257 | 0.2595285 | 0.1333076 |
| GB51159 | uncharacterized protein LOC413925 isoform X5 | 15.705241 | -0.4739860 | -0.2076484 | 0.2877617 | 0.1070991 |
| GB48933 | methenyltetrahydrofolate synthase domain-containing protein-like | 15.668023 | -0.8760827 | 0.0341808 | 0.5730859 | 0.1763404 |
| GB42606 | serotonin receptor | 15.665441 | -0.0399272 | -0.0611863 | 0.6977175 | 0.0109041 |
| GB43365 | nucleoside diphosphate kinase 7 | 15.646689 | -0.3570455 | -0.0098416 | 0.2813375 | 0.3381824 |
| 411207 | CCAAT/enhancer-binding protein | 15.645080 | -0.6195679 | -0.1924736 | 0.3585323 | -0.0524647 |
| 724740 | forkhead box protein G1 | 15.639002 | -0.2007863 | 0.2318166 | 0.6060152 | 0.1682673 |
| GB48451 | protein yippee-like 5-like isoform X1 | 15.604803 | 0.2249800 | 0.0831261 | 0.1174656 | 0.0902425 |
| GB46312 | cuticular protein 22 precursor | 15.571915 | 0.3941880 | -0.1897257 | 0.1996916 | -0.5429218 |
| GB40304 | ankyrin repeat and SOCS box protein 16-like isoform X2 | 15.562554 | -0.3703452 | -0.0217728 | 0.4131607 | -0.0338316 |
| GB40240 | myosin regulatory light chain 2 | 15.537435 | 0.5198008 | -0.4811814 | 0.2578078 | 0.2791373 |
| GB42875 | ATP-binding cassette sub-family A member 2-like isoformX2 | 15.495406 | -0.2022811 | -0.0643080 | 0.3013218 | -0.1625268 |
| GB41243 | uncharacterized protein LOC408729 isoform X2 | 15.481154 | -0.0961139 | -0.1463401 | 0.1604827 | -0.0858921 |
| GB47237 | protein naked cuticle homolog 2-like | 15.466641 | -1.4587881 | 0.2052271 | 0.3160004 | 0.0359239 |
| GB49400 | protein msta, isoform A-like isoform X3 | 15.436729 | 0.2130109 | 0.0692844 | 0.4102789 | 0.0730650 |
| 726815 | phosphatidylinositol-glycan biosynthesis class W protein-like | 15.405703 | -0.6211279 | -0.0432988 | 0.2534542 | 0.0559762 |
| GB44903 | calcineurin subunit B type 2-like | 15.400084 | -0.3690599 | -0.0323468 | 0.2586576 | 0.3400857 |
| GB40496 | aftiphilin-like isoform X2 | 15.353746 | 0.3123896 | -0.0021761 | 0.1516327 | 0.0668674 |
| GB40159 | ankyrin repeat and death domain-containing protein 1A-like isoform X3 | 15.338868 | -0.9287671 | -0.1708180 | 0.1862042 | -0.0916638 |
| GB47229 | heparan sulfate glucosamine 3-O-sulfotransferase 3A1 isoform X1 | 15.277497 | 0.7488732 | 0.1117085 | 0.4567357 | 0.0835861 |
| GB53055 | nicotinic acetylcholine receptor beta1 subunit precursor | 15.275694 | 0.8052158 | 0.6551395 | 0.3567407 | 0.4670622 |
| GB48208 | protein argonaute-2 isoform X4 | 15.254452 | -0.2092086 | -0.1481536 | 0.6297834 | 0.2431558 |
| 100576814 | uncharacterized protein LOC100576814 | 15.235960 | 0.3458361 | -0.2340662 | 1.1643477 | -0.1628239 |
| GB41745 | LOW QUALITY PROTEIN: serine/threonine-protein kinase atr-like | 15.163603 | -0.5220902 | 0.0056408 | -0.0634446 | 0.1541954 |
| GB41981 | jmjC domain-containing protein 4-like isoform 1 | 15.089675 | -0.6830191 | 0.0685019 | 0.2269399 | 0.1861268 |
| GB40416 | twist isoform X1 | 15.050220 | 0.1746768 | -0.1954057 | 0.3353427 | 0.0370247 |
| GB40545 | microprocessor complex subunit DGCR8-like isoform X4 | 15.014488 | -0.4215839 | 0.1644370 | 0.3061618 | 0.1654158 |
| GB47014 | wiskott-Aldrich syndrome protein family member 2-like isoform X2 | 14.992856 | -0.4790326 | 0.0490454 | 0.0720573 | -0.3700189 |
| GB42866 | bruchpilot | 14.984149 | -0.6732870 | -0.1107115 | 0.0068422 | 0.4366532 |
| GB52956 | synaptotagmin-10 | 14.956048 | 0.1403263 | -0.0496245 | 0.3017734 | 0.4816307 |
| GB45035 | histone acetyltransferase KAT6B-like | 14.950725 | 0.4555221 | -0.0375517 | 0.4085010 | 0.5484510 |
| 100578339 | condensin-2 complex subunit D3-like | 14.921732 | -0.9452916 | 0.0655228 | 0.3611837 | 0.0258053 |
| GB53550 | heat shock protein beta-1-like isoform X3 | 14.853049 | 0.1137667 | -0.1101359 | -0.0807538 | 0.2267810 |
| GB48698 | tetraspanin-7 isoform X1 | 14.850910 | 1.0420339 | 0.0949066 | 0.2067971 | 0.1425195 |
| GB55663 | LOW QUALITY PROTEIN: LIM/homeobox protein Lhx1-like | 14.789809 | -0.3226875 | 0.2402790 | 0.4224435 | -0.0745866 |
| GB51063 | uncharacterized protein LOC552276 isoform X1 | 14.736234 | -0.3592629 | -0.5973709 | 0.1580166 | 0.0362386 |
| GB48352 | TBC1 domain family member 14 | 14.723320 | -0.4388469 | 0.0523390 | 0.3386816 | -0.2360601 |
| GB42670 | WD repeat-containing protein 35-like | 14.692785 | 0.3135844 | 0.0031752 | 0.3421378 | -0.0070221 |
| GB49945 | BRCA1-associated RING domain protein 1-like isoform X1 | 14.683129 | -0.0553931 | -0.2586330 | 0.1972627 | 0.4190530 |
| GB44824 | corazonin receptor isoform X1 | 14.678732 | -0.3009799 | -0.1562374 | -0.1472673 | -0.1130528 |
| GB42576 | limbic system-associated membrane protein-like | 14.615751 | -0.6587715 | -0.0945517 | 0.2464464 | 0.2469177 |
| GB41082 | tyrosine-protein kinase Src42A-like isoform X5 | 14.596125 | 0.0388012 | 0.0337538 | 0.6878599 | 0.0900157 |
| GB43456 | 18-wheeler precursor | 14.595748 | -0.8609148 | 0.0679012 | -0.1989253 | 0.0263266 |
| GB53305 | LOW QUALITY PROTEIN: protein still life, isoform SIF type 1-like | 14.532129 | 0.1336125 | -0.1438707 | 0.6295653 | -0.0098067 |
| GB43389 | 8-oxo-dGDP phosphatase NUDT18-like | 14.494416 | 0.3072804 | -0.1161499 | 0.2413855 | -0.0762126 |
| GB49368 | rho-related GTP-binding protein RhoU-like | 14.451697 | 0.1988707 | -0.0075346 | 0.1519758 | 0.1489892 |
| GB49105 | ecdysteroid-regulated gene E74 isoform X10 | 14.440487 | -0.3805369 | -0.1026102 | 2.9210447 | 0.1422702 |
| GB44276 | discoidin domain-containing receptor 2-like | 14.362689 | 0.6563774 | -0.1394740 | 0.6697288 | 0.1013780 |
| GB49248 | tachykinins precursor | 14.345529 | 0.6788863 | -0.0240857 | 0.2638734 | 0.3006781 |
| GB49476 | lysyl oxidase homolog 4 isoform X2 | 14.253445 | -0.4140959 | -0.1441085 | 0.4441635 | 0.0050025 |
| GB41265 | nucleoredoxin-like isoform X3 | 14.208226 | -0.0250208 | 0.2309961 | 0.2964353 | 0.0450238 |
| GB50480 | mitochondrial inner membrane protein COX18-like isoform X1 | 14.196996 | -0.0499001 | -0.1484684 | 0.3402176 | 0.4398377 |
| GB50689 | L-threonine 3-dehydrogenase, mitochondrial-like | 14.181562 | 0.5938554 | -0.0330625 | 0.4041427 | 0.1300934 |
| 102653641 | glomulin-like | 14.154186 | -0.5667234 | -0.2011382 | 0.2555917 | 0.1108833 |
| GB54537 | protein slit isoform X2 | 14.075373 | -0.0126188 | -0.0843548 | 0.7197783 | 0.4252038 |
| GB43143 | tubulin-specific chaperone cofactor E-like protein-like isoform X2 | 14.023650 | 0.0612125 | -0.0809272 | 0.1838574 | -0.0942275 |
| GB51029 | band 4.1-like protein 5-like isoform X1 | 14.002161 | -0.6668539 | -0.0545778 | 0.0894735 | -0.0016594 |
| GB52719 | segmentation protein Runt-like | 13.759744 | -1.2877537 | 0.1757163 | 0.0618145 | -0.0312513 |
| GB47734 | uncharacterized protein LOC100577325 isoform 1 | 13.722415 | 0.2243826 | -0.0592934 | 0.2491272 | 0.0995916 |
| GB41085 | tektin-B1-like isoform X3 | 13.700821 | -0.3748334 | 1.0541514 | 0.5021390 | 0.4364121 |
| GB52002 | agrin-like isoform X10 | 13.648175 | -0.3814819 | 0.1502947 | 0.1656457 | 0.0423272 |
| GB53665 | tyramine beta hydroxylase | 13.554831 | -0.7343286 | -0.0328800 | 1.2794363 | 0.0629392 |
| 550870 | uncharacterized protein LOC550870 | 13.546847 | -1.8653555 | 0.3195073 | -0.0099437 | -0.0212492 |
| GB43719 | alpha-catulin-like isoform X2 | 13.486767 | -0.5041119 | 0.2381516 | 0.4204339 | 0.8312580 |
| GB44254 | uncharacterized protein LOC411586 isoform X1 | 13.354572 | 0.1548909 | -0.0759203 | 0.3807907 | 0.3661557 |
| GB54467 | probable G-protein coupled receptor 52 isoform 1 | 13.156538 | -1.0596453 | 0.1235973 | 0.4530378 | 0.3156867 |
| GB52328 | protein patched-like isoform X4 | 13.142650 | -0.0470780 | 0.0827657 | 0.4715143 | -0.1167653 |
| 102654980 | transient receptor potential channel pyrexia-like | 13.087750 | -0.5031178 | 0.0273209 | 0.6911742 | 0.1151823 |
| GB49844 | mediator of RNA polymerase II transcription subunit 1 isoform X1 | 13.038627 | -0.2415949 | -0.0343759 | 0.5057169 | -0.0666262 |
| GB51276 | protocadherin-like wing polarity protein stan-like isoform X1 | 12.994447 | -0.4409249 | -0.0213298 | 0.5249136 | -0.0773183 |
| GB43618 | aconitate hydratase, mitochondrial-like isoform X1 | 12.877769 | -0.4083154 | -0.1511920 | 0.1370423 | 0.1283512 |
| GB43292 | uncharacterized protein LOC551661 | 12.863419 | -0.6457428 | 0.2007350 | 0.3189085 | 0.2878090 |
| GB49175 | 4-hydroxyphenylpyruvate dioxygenase-like | 12.857044 | -0.3463731 | 0.3538868 | 0.5652253 | 0.1808402 |
| GB47373 | protein tipE-like isoform X1 | 12.800891 | -0.4067158 | 0.4340921 | 0.3845045 | 0.0012639 |
| 550677 | protein mab-21-like isoform 1 | 12.777109 | 0.3481217 | 0.2131949 | 0.3030159 | 0.0229235 |
| GB47370 | rho GTPase-activating protein 7 isoform X2 | 12.774984 | -0.3749962 | -0.0732343 | -0.4487794 | -0.3392253 |
| GB55974 | calcium uniporter protein, mitochondrial-like isoform X1 | 12.679886 | 0.3690429 | 0.1478188 | 0.2237683 | 0.3954706 |
| GB52995 | coiled-coil domain-containing protein 85C-like isoform X2 | 12.654932 | 0.5005453 | 0.1460103 | 0.3618843 | -0.1789676 |
| GB53420 | uncharacterized protein LOC100576355 isoformX2 | 12.616980 | -0.5907599 | 0.2092456 | 0.3775249 | -0.0155893 |
| GB40120 | fas apoptotic inhibitory molecule 1-like | 12.590428 | 0.3102156 | -0.5954096 | 0.0989003 | 0.3083215 |
| GB41601 | serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit A-like isoform 2 | 12.553174 | 0.2393149 | 0.0319892 | 0.2540168 | -0.2914843 |
| GB41227 | cuticular protein analogous to peritrophins 3-B precursor | 12.500511 | 0.1322101 | -0.6683104 | 0.4086942 | 0.0560315 |
| GB52992 | agrin-like | 12.495771 | 0.0225387 | -0.0731722 | 0.2459048 | -0.3041769 |
| GB53351 | 2-hydroxyacylsphingosine 1-beta-galactosyltransferase-like isoform 1 | 12.390049 | -0.6928586 | -0.0347413 | 1.0913462 | 0.3709605 |
| GB52356 | zeta-sarcoglycan isoformX2 | 12.329856 | 0.2223687 | -0.3003192 | 0.3488838 | -0.2510877 |
| GB42227 | homeobox protein aristaless-like | 12.322365 | 0.3371603 | 0.0445621 | -0.3980888 | 0.1229078 |
| GB47330 | uncharacterized protein LOC100578466 | 12.297337 | -0.1611251 | 0.2802525 | 2.1912450 | -0.0576892 |
| GB47902 | endocuticle structural glycoprotein SgAbd-1-like | 11.965446 | 0.9185689 | -0.3288476 | 0.2553881 | 0.4100066 |
| GB44295 | sodium-dependent neutral amino acid transporter B(0)AT2 isoform X1 | 11.869923 | -0.0389405 | 0.0150823 | 0.2602500 | 0.0721614 |
| GB51084 | radial spoke head protein 3 homolog isoform X1 | 11.741421 | 0.1292462 | -0.9700829 | 0.3011215 | 0.0656329 |
| GB47967 | TWiK family of potassium channels protein 7-like | 11.624651 | 0.2591543 | 0.1185417 | 0.5386654 | 0.3077002 |
| GB49882 | potassium voltage-gated channel protein Shaw isoform X2 | 11.395419 | -0.0336868 | -0.1443389 | 0.2748873 | 0.0978533 |
| GB45694 | lutropin-choriogonadotropic hormone receptor-like isoform X4 | 11.315345 | -0.3758967 | -0.2558261 | 1.2080970 | 0.4408211 |
| 100578156 | uncharacterized protein LOC100578156 | 11.077422 | -1.5700809 | 0.0780073 | 0.6243584 | 0.1110160 |
| GB41494 | uncharacterized protein LOC100576666 | 10.876470 | 0.2402189 | 0.1975217 | 1.0718525 | 0.3094964 |
| 409016 | uncharacterized protein LOC409016 isoform X1 | 10.234181 | 0.1886847 | -0.0454913 | 0.1942384 | 0.6256423 |
| GB48262 | uncharacterized LOC100576671, transcript variant X5 | 10.197905 | -0.6818715 | 0.0338790 | 0.0810053 | 0.0512313 |
| GB51174 | uncharacterized protein DDB_G0284459-like | 10.039886 | 0.4280375 | -0.3750459 | 0.1868244 | 0.5932720 |
| GB46638 | dipeptidase 1-like | 9.993053 | -0.2757522 | -0.7356442 | -0.4211237 | 0.3103322 |
| GB46057 | PBAN-type neuropeptides precursor | 9.971053 | 0.0272047 | -0.4366709 | 0.1581147 | 0.1132313 |
| GB55016 | quinone oxidoreductase-like isoform X2 | 9.863561 | 0.6216726 | -0.0803903 | 0.4652785 | -0.1927008 |
| GB49392 | actin-binding Rho-activating protein-like isoform 1 | 9.259932 | 0.7867372 | -0.1883898 | 0.3315127 | 0.1847873 |
| GB48832 | cuticular protein 3 precursor | 9.092388 | 1.5395500 | -0.1942970 | 0.3957273 | 0.1781706 |
| GB54396 | elongation of very long chain fatty acids protein AAEL008004-like isoform X2 | 8.959702 | 2.4882823 | -0.0852643 | 0.9730720 | 0.0513045 |
| GB46335 | uncharacterized protein LOC100577622 | 8.870681 | 0.2761928 | 0.1877337 | 1.1725539 | 0.0700643 |
| GB45199 | ceramide kinase-like isoform X4 | 8.782776 | 0.2247593 | 0.0685426 | 0.1886628 | -0.1044807 |
| 724465 | enhancer of split mbeta protein-like | 8.744289 | 1.0913239 | 0.0051604 | 0.6081269 | -0.1332904 |
| GB41903 | cyclin N-terminal domain-containing protein 1-like isoform X2 | 8.419051 | -0.1875021 | 0.4823907 | 0.4797190 | 0.7172901 |
| 724570 | mpv17-like protein 2-like isoform X2 | 8.120352 | 0.2675164 | -0.0182479 | 0.1683108 | 0.1254560 |
| GB44549 | glucose oxidase | 7.627876 | 0.0464020 | 0.4531259 | 0.1390218 | -0.5851793 |
| GB47092 | uncharacterized protein LOC724483 isoform X1 | 7.416226 | -0.5250113 | 0.7248697 | 0.4849716 | -0.0539099 |
| GB50889 | RNA-binding protein 24-like isoform X3 | 7.323361 | -0.0343137 | 0.0133994 | 0.6501237 | 0.1515999 |
| GB43560 | insulin-like peptide 2 | 7.254103 | 0.3891473 | -0.1541425 | 0.2881154 | 0.2171360 |
| GB51740 | CD63 antigen | 7.148564 | 0.2998193 | -0.1110492 | 2.6283593 | 0.0933423 |
| GB49684 | optomotor-blind protein-like isoform X1 | 7.087644 | 0.1104555 | 0.1842527 | 0.0932658 | -0.1836892 |
| GB45542 | ligand-gated chloride channel homolog 3 precursor | 6.964167 | 1.9871990 | 0.1979414 | 0.9314630 | 0.4740336 |
| GB51369 | opsin, ultraviolet-sensitive | 6.544346 | 0.5283196 | 0.0686868 | 0.1735806 | 0.2177808 |
| GB45157 | protein big brother isoform 1 | 5.982996 | -0.1646934 | 0.2312355 | 0.2537482 | -0.0448091 |
| GB13601 | cuticular protein CPF1 precursor | 5.918821 | 0.5209523 | -0.1661667 | 0.3437081 | 0.1136129 |
| GB41418 | uncharacterized protein PF11_0207-like isoform X2 | 5.728419 | 0.6725455 | 0.0729602 | 0.1688289 | 0.7614956 |
| GB47727 | uncharacterized protein LOC724679 isoform 2 | 4.795946 | 0.5902646 | -0.1623603 | 0.3408201 | 0.0704282 |
| 102656656 | carbon catabolite-derepressing protein kinase-like isoform X2 | 4.511399 | -0.2794717 | -0.0120319 | 0.4746257 | 0.0482806 |
| GB54159 | uncharacterized protein LOC552735 | 4.176112 | 0.3288372 | 0.1544875 | 0.3665780 | 0.3262821 |
Module 4
Table S29: List of all the genes in Module 4, ranked by their within-module connectivity, k. The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
gene_list <- module_gene_list(4)
saveRDS(gene_list, file = "supplement/tab_S29.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB44431 | 26S protease regulatory subunit 4 isoform 1 | 44.223556 | 0.2068368 | -0.0420157 | -0.4280960 | -0.0088204 |
| GB49337 | 26S proteasome non-ATPase regulatory subunit 13 | 42.984225 | 0.1764581 | -0.0655698 | -0.2607125 | -0.5605728 |
| GB50750 | coatomer subunit delta isoform 2 | 42.890303 | -0.0312791 | -0.0288217 | -0.0588197 | -0.0489833 |
| GB45720 | proteasome subunit alpha type-7-1-like | 41.581027 | 0.1530987 | -0.0949419 | -0.0722067 | 0.0630901 |
| GB50242 | 26S proteasome non-ATPase regulatory subunit 4 | 41.384303 | 0.2213651 | 0.0631539 | -0.1328648 | 0.5944635 |
| GB53174 | programmed cell death 6-interacting protein isoform X1 | 41.242520 | -0.0756447 | -0.0159918 | -0.0466398 | -0.0545396 |
| GB47189 | H(+)/Cl(-) exchange transporter 3-like isoform X2 | 41.034107 | 0.0346407 | -0.0154327 | -0.1018644 | -0.0454175 |
| GB50252 | GTP-binding protein SAR1b-like isoform X4 | 40.648326 | 0.3208936 | -0.0667845 | -0.0210473 | -0.2223932 |
| 102656372 | tropinone reductase 2-like | 39.868937 | 0.1120373 | 0.0340800 | 0.3238888 | 0.4926542 |
| GB53812 | prolyl endopeptidase-like isoformX1 | 39.532524 | -0.0804557 | -0.0489813 | -0.0926730 | -0.0238204 |
| GB45567 | E3 ubiquitin-protein ligase MARCH5 isoform X5 | 39.470727 | -0.0551062 | 0.0346992 | 0.4749971 | 0.0134325 |
| GB41649 | E3 ubiquitin-protein ligase MARCH6 | 39.278828 | -0.0462292 | -0.0638063 | -0.0682451 | 0.0497685 |
| GB43819 | ATPase ASNA1 homolog | 39.180343 | 0.3773737 | -0.0065874 | -0.1431066 | -0.0100659 |
| GB44941 | eukaryotic peptide chain release factor GTP-binding subunit ERF3A | 39.104220 | -0.0593707 | 0.0722754 | -0.0148940 | -0.1104174 |
| GB53690 | protein transport protein Sec31A | 38.707647 | -0.0073805 | 0.0306890 | -0.1181363 | -0.0320893 |
| GB48643 | STT3, subunit of the oligosaccharyltransferase complex, homolog B | 37.962742 | 0.1934304 | 0.0005876 | -0.1604664 | -0.0954778 |
| GB50348 | bleomycin hydrolase-like isoform X2 | 37.858985 | -0.0331317 | 0.0361318 | -0.0548069 | 0.0615163 |
| GB43706 | probable trans-2-enoyl-CoA reductase, mitochondrial-like | 37.473883 | -0.0105284 | 0.0051152 | -0.0134324 | 0.0351066 |
| GB40775 | apoptosis-inducing factor 1, mitochondrial | 36.869445 | 0.2000561 | 0.1909935 | 0.3170325 | 0.0325652 |
| 411789 | protein extra bases | 36.857043 | 0.1887237 | 0.0207032 | -0.1285144 | 0.1008955 |
| GB40483 | xyloside xylosyltransferase 1-like | 36.852931 | 0.0301872 | 0.0061033 | -0.0806551 | 0.0684976 |
| GB41617 | methylcrotonoyl-CoA carboxylase beta chain, mitochondrial-like isoform X2 | 36.788246 | 0.1487599 | -0.1687086 | -0.2389065 | -0.0591055 |
| GB50289 | signal recognition particle 54 kDa protein-like | 36.640755 | 0.0039203 | -0.0010764 | -0.0138630 | -0.0199337 |
| GB50274 | transitional endoplasmic reticulum ATPase TER94 | 36.259304 | 0.5435617 | -0.1180270 | -0.3064504 | -0.0779525 |
| GB47540 | putative leucine-rich repeat-containing protein DDB_G0290503 isoform X2 | 36.214750 | 0.2775390 | 0.0232034 | -0.1293343 | 0.0217957 |
| GB47573 | T-complex protein 1 subunit theta-like | 36.082403 | 0.1562527 | 0.0513066 | -0.0207824 | -0.0623897 |
| GB52675 | 26S proteasome non-ATPase regulatory subunit 12 | 35.929513 | 0.2312931 | 0.0482900 | -0.0951427 | 0.7829533 |
| GB42355 | asparagine–tRNA ligase, cytoplasmic-like | 35.911929 | 0.1286098 | 0.1015732 | -0.1220399 | -0.0430047 |
| GB42329 | segmentation protein cap’n’collar-like isoform X4 | 35.867197 | -0.2192666 | -0.1006952 | -0.1213930 | 0.6014040 |
| GB55494 | probable nucleolar GTP-binding protein 1-like isoform 1 | 35.759099 | 0.0447120 | 0.0957711 | 3.8215256 | -0.1220854 |
| GB42773 | alanine–tRNA ligase, cytoplasmic-like isoform X1 | 35.229118 | -0.0350767 | 0.0415367 | -0.0868974 | 0.0119881 |
| GB50730 | 97 kDa heat shock protein isoformX1 | 35.063680 | -0.1577197 | -0.0288468 | -0.2107992 | -0.1611854 |
| GB48313 | transmembrane 9 superfamily member 3 | 34.685061 | 0.2534914 | -0.0136668 | -0.0312486 | -0.0353920 |
| GB50459 | WD repeat-containing protein 36-like | 34.578669 | -0.1479411 | -0.0430390 | -0.2433271 | -0.1232282 |
| GB46035 | eukaryotic translation initiation factor 4 gamma 2-like isoform X4 | 33.921974 | -0.2214407 | 0.0295406 | -0.2425567 | -0.1486897 |
| GB54608 | probable elongator complex protein 2-like | 33.814252 | 0.0689425 | 0.0691345 | -0.2205388 | -0.1308013 |
| GB50177 | protein TRC8 homolog | 33.492549 | 0.0945129 | 0.1231066 | -0.1733043 | 0.2465639 |
| GB45258 | isocitrate dehydrogenase [NADP] cytoplasmic isoform 2 | 33.420505 | -0.0142164 | -0.0157218 | -0.1189860 | -0.0987758 |
| GB47114 | dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2-like | 33.136515 | 0.1350366 | 0.1040145 | -0.1152653 | -0.1108689 |
| GB44418 | protein suppressor of hairy wing isoform X2 | 33.079975 | -0.4107368 | -0.1260784 | -0.2722544 | 0.0277971 |
| GB54573 | probable 26S proteasome non-ATPase regulatory subunit 3 isoform X2 | 31.928635 | 0.0538327 | -0.0037021 | -0.0681125 | -0.9049955 |
| GB55440 | phosphatidylinositol transfer protein alpha isoform | 31.910100 | 0.1654650 | 0.0068526 | -0.0463461 | 0.0630027 |
| GB41762 | derlin-2-like | 31.704673 | 0.1842082 | 0.0872893 | -0.2212623 | -0.1243794 |
| GB49939 | protein FAM188A homolog | 31.551690 | -0.1412746 | 0.1136928 | -0.1907462 | 0.0388591 |
| GB42648 | dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 | 31.480759 | 0.2655380 | 0.1595473 | -0.1227576 | -0.1662744 |
| GB44670 | luciferin 4-monooxygenase-like | 31.430736 | -0.0515925 | 0.1548945 | 1.1032901 | 0.0559438 |
| GB43131 | short-chain dehydrogenase/reductase family 16C member 6-like isoform X4 | 31.410234 | 0.1007306 | 0.0869099 | -0.1985410 | -0.0829191 |
| GB48692 | translocation protein SEC63 homolog isoform 1 | 31.125908 | 0.2857900 | -0.0530291 | -0.1602624 | 0.0209032 |
| GB49083 | casein kinase II subunit beta isoform X1 | 30.992393 | 0.2782502 | 0.0471649 | -0.1111348 | 0.0698090 |
| GB41427 | catalase | 30.732896 | 0.1430821 | 0.0736901 | -0.3977072 | -0.0121187 |
| GB46735 | eukaryotic translation initiation factor 2A-like | 30.705422 | -0.0411662 | 0.0130254 | -0.1447461 | -0.0105182 |
| GB47296 | transmembrane protein 19-like isoform X2 | 30.672830 | 0.4289691 | -0.0545500 | -0.2516466 | -0.1027988 |
| GB52168 | von Willebrand factor A domain-containing protein 8-like | 30.577444 | 0.0856555 | -0.0740676 | 0.0014012 | 0.0699640 |
| GB48812 | dnaJ homolog subfamily C member 3 | 30.546137 | 0.1132367 | 0.0402074 | -0.2195332 | 0.0306329 |
| 102655967 | ancient ubiquitous protein 1-like | 30.348262 | 0.4711330 | 0.0420199 | 0.1648789 | 0.0503451 |
| 551499 | FIT family protein CG10671-like | 29.740675 | 0.3785333 | -0.0602509 | -0.3180826 | -0.0349339 |
| GB49955 | vacuole membrane protein 1 isoform X1 | 29.459325 | -0.0443525 | -0.0065202 | -0.2963167 | 0.1983572 |
| GB41285 | receptor-binding cancer antigen expressed on SiSo cells | 29.433269 | 0.0238298 | 0.1200911 | -0.1399244 | -0.1309690 |
| GB54861 | LOW QUALITY PROTEIN: counting factor associated protein D-like | 29.320266 | 0.2298241 | 0.1926641 | -0.2944184 | -0.0974716 |
| GB47462 | protein disulfide-isomerase A3 isoform 2 | 29.268674 | 0.3049034 | 0.0771835 | -0.2557549 | 0.1369471 |
| GB46646 | UPF0554 protein C2orf43 homolog | 28.707580 | 0.3354412 | -0.0495704 | -0.2178741 | -0.1430792 |
| GB44396 | atlastin isoform X2 | 28.556709 | -0.0591485 | -0.1777902 | -0.0925969 | 0.3200035 |
| GB53373 | leucine-rich repeat-containing protein 58-like | 28.367576 | 0.1837683 | 0.0574237 | -0.2601640 | 0.1579574 |
| GB55484 | UBX domain-containing protein 7-like | 28.345099 | -0.1419776 | 0.0862990 | -0.1770521 | -0.0476737 |
| GB47134 | renin receptor-like isoform X1 | 28.303254 | 0.3303687 | -0.0407767 | 0.3718194 | 0.4166502 |
| GB43912 | nicalin-1 isoform X1 | 28.248461 | 0.6287100 | 0.0881076 | -0.3253488 | -0.0103644 |
| GB46120 | aspartate aminotransferase, mitochondrial isoform 1 | 28.225409 | 0.2491141 | 0.0236496 | 0.1306639 | 0.0254197 |
| GB49525 | RNA-binding protein fusilli | 28.209030 | 0.1174438 | -0.0741709 | -0.0349456 | 0.0534866 |
| GB48597 | NADH-cytochrome b5 reductase 2-like isoform X2 | 28.155088 | 0.2033803 | 0.0013862 | -0.0996939 | -0.0202651 |
| GB44205 | proteasome subunit beta type-5-like | 28.006936 | 0.4837999 | -0.0915736 | -0.4335090 | -0.0596112 |
| GB52434 | probable ribosome production factor 1-like | 27.874103 | 0.0054318 | 0.0148853 | -0.2060277 | 0.0182433 |
| GB40779 | transaldolase | 27.760521 | -0.0039251 | 0.0160977 | -0.0440904 | 0.0344095 |
| GB45582 | facilitated trehalose transporter Tret1-like isoform 3 | 27.374644 | -0.0504268 | -0.1335269 | 0.0539026 | -0.0069750 |
| GB45698 | SAGA-associated factor 11 homolog | 27.343826 | -0.2647791 | 0.0924924 | -0.1384764 | 0.1879171 |
| GB40265 | transcriptional activator protein Pur-beta-B-like isoform X4 | 27.334314 | 0.1264999 | -0.0386187 | -0.1285937 | -0.0782933 |
| GB47392 | protein BCCIP homolog | 27.258892 | 0.2577727 | 0.1104351 | 0.7443079 | 0.1332378 |
| GB55987 | ras-related protein Rab-18-B | 27.110725 | 0.0323400 | -0.0536913 | -0.0349277 | -0.0504278 |
| GB55977 | eukaryotic translation initiation factor 4E-1A | 27.089946 | 0.1705188 | 0.0538447 | -0.0949449 | 0.1031615 |
| GB45251 | ubiA prenyltransferase domain-containing protein 1 homolog | 26.995162 | 0.2292889 | 0.0114163 | -0.1159756 | 0.0793336 |
| GB48008 | peroxisomal membrane protein PEX14-like | 26.969623 | 0.0001390 | 0.0224702 | -0.2071820 | -0.0800297 |
| GB46977 | ribosome biogenesis methyltransferase WBSCR22-like | 26.952365 | 0.0481658 | 0.0896746 | -0.2062109 | 0.0893196 |
| GB54363 | ATPase family AAA domain-containing protein 3 isoform X1 | 26.942276 | 0.3943176 | 0.1631272 | -0.1500874 | -0.0561398 |
| GB53955 | FGFR1 oncogene partner 2 homolog | 26.809933 | 0.0846268 | -0.0312388 | -0.1442031 | 0.0964404 |
| GB51282 | thioredoxin domain-containing protein 5-like isoform 1 | 26.691792 | 0.2485396 | 0.0763851 | 0.1463655 | -0.0113567 |
| GB46031 | vacuolar H+ ATP synthase 16 kDa proteolipid subunit | 26.645728 | -0.0479109 | -0.0170686 | -0.2826372 | 0.0222715 |
| GB54848 | tumor suppressor candidate 3-like | 26.523213 | 0.4846324 | -0.0104631 | -0.6272344 | -0.0006124 |
| GB49119 | deoxynucleotidyltransferase terminal-interacting protein 2-like | 26.458317 | 0.2109767 | -0.0144902 | -0.1065953 | 0.0599639 |
| GB53080 | alpha-2-macroglobulin receptor-associated protein-like | 26.455789 | 0.0064170 | -0.0306365 | -0.0869340 | 0.0369668 |
| GB49117 | heat shock protein cognate 3 precursor | 26.360858 | 0.2270221 | -0.0395375 | -0.2932567 | -0.1580736 |
| GB49240 | aldehyde dehydrogenase, mitochondrial isoform 1 | 26.349079 | 0.1549590 | 0.1010275 | -0.0376052 | -0.0094109 |
| GB55610 | MOSC domain-containing protein 2, mitochondrial-like | 26.339169 | -0.0627471 | 0.0072092 | 0.3240534 | -0.1399329 |
| GB49307 | DNA-directed RNA polymerases I and III subunit RPAC1-like isoform X1 | 26.293649 | -0.0115285 | 0.0668726 | 0.3393313 | -0.0367022 |
| GB52729 | aspartate–tRNA ligase, cytoplasmic | 26.270157 | 0.1271826 | -0.0104019 | 7.4806151 | -0.0363266 |
| GB40207 | serine–tRNA ligase, mitochondrial | 26.269427 | 0.0012962 | -0.0721504 | 0.0021323 | -0.0446679 |
| GB46979 | derlin-1-like | 26.162259 | 0.6809454 | -0.0344909 | 0.5101016 | -0.0139263 |
| GB42236 | patched domain-containing protein 3-like isoform X4 | 25.938301 | -0.2888681 | 0.0901237 | -0.1149775 | 0.1142936 |
| 102653839 | histone-lysine N-methyltransferase SETMAR-like | 25.496262 | 0.2063172 | -0.0636157 | -0.2265030 | -0.1742015 |
| GB49180 | cysteine-rich secretory protein 1-like, transcript variant X5 | 25.371025 | -0.2540170 | -0.0393153 | -0.1190649 | -0.2517126 |
| GB55537 | transketolase isoform 1 | 25.321298 | 1.0357857 | 0.0369899 | -0.2479617 | -0.3417730 |
| GB54999 | NAD kinase 2, mitochondrial-like | 25.209366 | 0.4540230 | -0.2140000 | -0.0815604 | -0.0922070 |
| GB54101 | HEAT repeat-containing protein 3-like | 24.883603 | -0.2253708 | -0.0872323 | -0.1050610 | 0.0597797 |
| GB55490 | uncharacterized protein LOC410793 | 24.753469 | -0.0477751 | -0.0681408 | -0.4730765 | 0.2615252 |
| GB46579 | glucose-6-phosphate 1-dehydrogenase isoform X3 | 24.688398 | 0.6482683 | -0.1000294 | -0.3650464 | -0.1593256 |
| GB50096 | pantothenate kinase 1-like isoform X2 | 24.586554 | 0.8477323 | -0.0870535 | -0.3693640 | -0.0267439 |
| GB54112 | adenine phosphoribosyltransferase isoform X1 | 24.536162 | 0.2345084 | 0.0448087 | 1.4763523 | 0.1039490 |
| GB47432 | 5-aminolevulinate synthase, erythroid-specific, mitochondrial-like | 24.361288 | 0.3065397 | -0.2485980 | -0.3612845 | -0.0388571 |
| GB40783 | glucose-6-phosphate isomerase-like | 24.207603 | -0.2439304 | -0.1024763 | -0.0910389 | -0.0013179 |
| GB54298 | stromal cell-derived factor 2-like protein 1-like isoformX2 | 24.138324 | 0.3180715 | 0.0516537 | -0.2166825 | 1.5192543 |
| GB44457 | FGGY carbohydrate kinase domain-containing protein-like isoform X2 | 23.955079 | 0.3022943 | -0.1605569 | -0.2354506 | -0.0710812 |
| GB48408 | protein catecholamines up | 23.947718 | 0.0141547 | -0.0116125 | -0.1088589 | 0.0730103 |
| GB48847 | DNA replication licensing factor Mcm3 | 23.786913 | -0.3084852 | 0.2500155 | -0.2998054 | 0.1204342 |
| GB46657 | galactokinase-like | 23.579329 | 0.1002480 | 0.0684625 | -0.5178679 | 0.0198896 |
| GB52347 | saccharopine dehydrogenase-like oxidoreductase-like isoform 1 | 23.401023 | 0.3645361 | -0.4951154 | 0.2533578 | -0.1998256 |
| GB51782 | carboxypeptidase Q-like isoform 1 | 23.387758 | 0.3346236 | -0.0203412 | -0.0649589 | -0.1664672 |
| GB48308 | probable pyruvate dehydrogenase E1 component subunit alpha, mitochondrial-like isoform X2 | 23.311737 | -0.2837578 | -0.1078497 | 0.1711523 | 0.5184012 |
| GB44557 | probable ribonuclease ZC3H12C-like isoformX1 | 23.279307 | 0.0744929 | -0.0447778 | -0.1440084 | -0.3265611 |
| GB47941 | cyclic AMP response element-binding protein A-like | 23.225979 | -0.3397125 | 0.1288849 | -0.1450569 | -0.3071147 |
| GB42732 | long-chain-fatty-acid–CoA ligase 3-like isoform X2 | 23.218782 | 0.4989058 | -0.0146724 | -0.3252347 | -0.1571386 |
| GB52724 | protein 5NUC-like isoform X2 | 23.028043 | 0.0972367 | 0.0206395 | -0.1179516 | 0.0023551 |
| GB46772 | very-long-chain enoyl-CoA reductase-like | 23.018130 | 0.7467499 | -0.2199431 | -0.2011022 | -0.4308688 |
| GB44008 | BTB/POZ domain-containing protein 17 isoform X1 | 22.839645 | 0.3154921 | 0.1167502 | -0.1116056 | -0.0483891 |
| GB55511 | growth/differentiation factor 8-like isoform 1 | 22.816925 | 0.4352292 | 0.1588802 | -0.4870545 | 0.0743345 |
| GB54601 | protein disulfide-isomerase A6-like isoform 1 | 22.775763 | 0.4324571 | 0.1249594 | -0.1467693 | -0.0779340 |
| GB49342 | sugar phosphate exchanger 2-like isoform X3 | 22.718190 | -0.2236853 | -0.0107231 | -0.0067022 | 0.0247381 |
| GB49348 | transmembrane protein 115-like | 22.714857 | 0.3394745 | -0.0382297 | -0.0306424 | -0.0288120 |
| GB41388 | glycerol-3-phosphate dehydrogenase | 22.668744 | -0.0851797 | -0.1391961 | -0.1883058 | 0.2562985 |
| GB49336 | acetyl-CoA carboxylase-like isoform X9 | 22.667437 | 0.3952476 | -0.1602047 | 0.1869465 | -0.2268771 |
| GB54056 | serine hydroxymethyltransferase, cytosolic isoform X3 | 22.549057 | 0.4140579 | 0.1933755 | -0.3256797 | -0.2607203 |
| GB44640 | solute carrier family 52, riboflavin transporter, member 3-A-like isoform X2 | 22.453147 | -0.1275523 | -0.1027834 | 0.1884364 | -0.2170403 |
| GB49826 | sterol O-acyltransferase 1-like | 22.192329 | 0.0951372 | 0.1465247 | -0.2836459 | 0.0229186 |
| GB47694 | globin 1 | 22.014317 | -0.0509277 | 0.1091362 | -0.1515248 | -0.1010404 |
| GB52074 | 6-phosphogluconate dehydrogenase, decarboxylating | 21.966722 | 0.8095794 | -0.1426001 | -0.4446501 | -0.6031632 |
| GB48195 | acyl-CoA Delta(11) desaturase-like | 21.937703 | 0.4943072 | -0.1931849 | 0.6995588 | 0.6686102 |
| GB45213 | acyl-CoA synthetase short-chain family member 3, mitochondrial-like isoform X2 | 21.833726 | 0.6513876 | 0.2239474 | -0.5637999 | -0.2397701 |
| GB50680 | mannose-P-dolichol utilization defect 1 protein homolog isoform X2 | 21.555807 | 0.4656771 | -0.3933720 | -0.2207091 | -0.0486393 |
| GB45775 | pancreatic triacylglycerol lipase-like isoform X2 | 21.506443 | 0.3197015 | 0.0505266 | -0.8880584 | 0.0691686 |
| GB41916 | uncharacterized protein LOC726658 isoform 1 | 21.370720 | 0.3361149 | 0.3728404 | -0.0276391 | -0.0165127 |
| GB52458 | cysteine-rich with EGF-like domain protein 2-like | 21.264629 | -0.2710389 | 0.0498794 | -0.3606772 | -0.0178730 |
| GB40278 | probable methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial isoform X4 | 21.241766 | -0.1582318 | 0.0029943 | 0.0294265 | -0.0342272 |
| GB54216 | ATP-citrate synthase isoform X2 | 21.223912 | 0.8455868 | -0.2267974 | -0.4457293 | -0.3264580 |
| 552211 | protein THEM6-like | 21.184585 | 0.7375726 | -0.1442659 | 0.3923799 | -0.5969306 |
| GB55533 | RNA-binding protein squid-like | 20.856600 | 0.0651253 | 0.0778979 | -0.0906335 | -0.1167986 |
| GB48859 | UPF0160 protein MYG1, mitochondrial-like isoform X2 | 20.818643 | 0.2361138 | 0.2491324 | -0.2080104 | -0.0332518 |
| GB49433 | H/ACA ribonucleoprotein complex subunit 2-like protein | 20.790828 | 0.5517963 | 0.1195184 | -0.1634197 | 0.0024375 |
| GB42237 | N6-adenosine-methyltransferase 70 kDa subunit-like | 20.731214 | -0.0894263 | 0.2756185 | -0.0857468 | -0.0529242 |
| GB46921 | monocarboxylate transporter 12-like | 20.714713 | 0.1807485 | 0.0007541 | -0.1003586 | -0.2148760 |
| GB45596 | elongation of very long chain fatty acids protein 6-like | 20.705352 | 0.5247867 | -0.4973462 | -0.5304114 | -0.5077637 |
| GB50013 | proclotting enzyme | 20.601511 | 0.2512238 | -0.9767295 | -0.5331162 | -0.2907466 |
| GB55263 | putative fatty acyl-CoA reductase CG5065-like | 20.420422 | -0.0329414 | -0.2619832 | -0.8318867 | -0.3621781 |
| GB54404 | elongation of very long chain fatty acids protein AAEL008004-like | 20.311401 | 2.0337166 | -0.0397052 | -0.1819294 | -0.1518170 |
| GB55094 | protein neuralized isoform X3 | 20.264296 | -0.6029457 | 0.1077662 | -0.6403894 | -0.0243416 |
| GB54427 | ribonucleoside-diphosphate reductase subunit M2 isoform X2 | 20.130036 | 0.0040836 | 0.1604850 | -0.0329101 | 0.0177491 |
| GB54538 | uncharacterized protein LOC411248 isoform X5 | 20.086236 | -0.4660266 | -0.2286794 | -0.0025983 | -0.0413626 |
| GB52768 | alkaline phosphatase, tissue-nonspecific isozyme-like isoform X1 | 20.080226 | -0.0170971 | 0.1366385 | -0.0403085 | -0.0385024 |
| GB51580 | long-chain-fatty-acid–CoA ligase ACSBG2 isoform X1 | 20.069108 | -0.0990216 | -0.1748159 | -0.2690449 | -0.1224712 |
| GB50871 | serine/threonine-protein kinase SIK2-like isoform X2 | 19.916686 | 0.0606616 | -0.1441363 | -0.2312372 | 0.2129038 |
| GB53287 | sialin-like isoform X4 | 19.620220 | -0.0297581 | -0.1059771 | -0.1192229 | -0.2783113 |
| GB49653 | probable phosphoserine aminotransferase-like | 19.478348 | 0.4313976 | 0.0935267 | 0.0591799 | -0.0947978 |
| GB47495 | nucleotide exchange factor SIL1-like | 19.344565 | -0.1521038 | 0.1578777 | -0.0097726 | -0.0068981 |
| GB51723 | 60S ribosomal export protein NMD3 | 19.222509 | -0.0712135 | 0.1664404 | 0.1759210 | -0.0663748 |
| GB48628 | RNA-binding protein Nova-1-like isoform X2 | 19.214104 | 0.0515613 | 0.0209321 | -0.5161615 | -0.1714978 |
| GB50626 | phospholipase D3-like isoform X7 | 19.160241 | -0.1767844 | -0.0946056 | -0.1617004 | -0.0368420 |
| GB54331 | cathepsin L-like isoform X2 | 19.064368 | 0.0167944 | -0.0794017 | -0.3784244 | -0.0640108 |
| GB53412 | fatty acid synthase-like | 18.867764 | 1.1288740 | -0.3170657 | 0.5061635 | -0.1800755 |
| GB51753 | uncharacterized protein LOC100576760 isoform X2 | 18.820612 | 0.0773965 | 0.1404132 | -0.0662854 | -0.1625608 |
| 100577899 | DNA replication complex GINS protein SLD5-like | 18.733489 | 0.3469362 | 0.1156921 | -0.2384693 | -0.0444013 |
| GB42899 | uncharacterized protein LOC551133 isoform X2 | 18.387282 | 0.1289445 | 0.0095466 | -0.0099508 | -0.0564785 |
| GB52446 | uncharacterized protein LOC726987 isoform X5 | 18.323554 | -0.5188239 | -0.2066036 | -0.3780834 | -0.1773838 |
| GB52351 | porphobilinogen deaminase-like | 18.239714 | -0.3935509 | -0.0800125 | -0.5113538 | 0.0303557 |
| GB45381 | putative sodium-coupled neutral amino acid transporter 7-like | 18.232994 | 0.2322221 | -0.2991610 | -0.1452851 | -0.1044424 |
| GB41886 | protein transport protein Sec61 subunit alpha isoform 2 | 18.215808 | 0.7287917 | -0.0327566 | -0.2604661 | -0.3382911 |
| GB52153 | U3 small nucleolar RNA-associated protein 15 homolog | 18.207361 | 0.1742820 | 0.1472587 | -0.0709591 | -0.0205023 |
| GB48203 | laminin subunit beta-1 isoform X2 | 18.135401 | -0.0687494 | 0.0552598 | -0.1815586 | -0.5195985 |
| GB51647 | 4-aminobutyrate aminotransferase, mitochondrial-like isoform X2 | 18.132631 | 0.5676800 | 0.0956367 | -0.1094744 | -0.0314973 |
| GB52454 | mitochondrial pyruvate carrier 2-like | 18.031851 | -0.4590473 | 0.0544465 | -0.1848845 | 0.0310854 |
| GB49942 | mitochondrial dicarboxylate carrier-like isoform 1 | 17.737991 | 0.1890512 | -0.0364015 | -0.1444554 | -0.0878453 |
| GB51614 | probable methylthioribulose-1-phosphate dehydratase-like | 17.686800 | -0.0358269 | -0.0730244 | 2.4507128 | 0.0926130 |
| GB41011 | lateral signaling target protein 2 homolog | 17.643002 | -0.0761978 | -0.1736913 | -0.5037217 | -0.0906862 |
| GB49869 | microsomal triglyceride transfer protein large subunit isoform X1 | 17.589998 | 0.1389107 | 0.2371633 | -0.4710017 | -0.8792260 |
| GB55432 | glucosidase 2 subunit beta-like | 17.428805 | 0.4916918 | 0.1151030 | -0.1781667 | 0.0426794 |
| GB40071 | uncharacterized protein LOC410446 | 17.398408 | 0.0764242 | 0.0301336 | 1.2168758 | 0.0847128 |
| GB44888 | MATH and LRR domain-containing protein PFE0570w-like | 17.235171 | 0.0429701 | -0.0248396 | 0.0078619 | -0.1299584 |
| GB54610 | thiamine transporter 2-like, transcript variant X2 | 17.225455 | -0.4767433 | 0.1088769 | -0.1870434 | -0.3209313 |
| GB54661 | phosphoglucomutase isoform X2 | 17.097639 | -0.3363439 | -0.1135590 | -0.1806090 | -0.2087559 |
| GB46422 | proton-coupled amino acid transporter 1 | 16.930428 | 0.2093788 | -0.1884596 | 0.0117507 | -0.1665571 |
| GB45177 | uncharacterized protein LOC725324 isoform X1 | 16.879588 | 0.4068916 | -0.0574424 | -0.1430491 | 0.3385230 |
| GB49633 | RNA 3’-terminal phosphate cyclase-like protein-like isoform X2 | 16.859461 | 0.1575109 | -0.0608420 | -0.0908976 | 0.0954425 |
| GB40141 | venom serine carboxypeptidase | 16.809434 | 0.1223739 | -0.3228364 | -0.3209092 | -0.0884225 |
| GB40280 | pyruvate carboxylase, mitochondrial isoform X1 | 16.742568 | -0.6020803 | -0.0510597 | -0.2153246 | -0.3460215 |
| GB49757 | fatty acid binding protein | 16.704757 | 0.4148758 | -0.1762910 | 0.3588453 | -0.1569230 |
| GB46661 | sodium-independent sulfate anion transporter-like isoform X1 | 16.610634 | -0.2846534 | 0.2134063 | -0.0536858 | -0.0012242 |
| GB45210 | translocon-associated protein subunit gamma-like | 16.589367 | -0.2578927 | -0.0623966 | -0.0363309 | -0.1380282 |
| GB47383 | U4/U6 small nuclear ribonucleoprotein Prp4 | 16.525971 | 0.0183237 | -0.0212914 | -0.0394002 | 0.0668049 |
| GB42787 | dentin sialophosphoprotein-like isoform X4 | 16.515172 | 0.1684747 | 0.3042534 | -0.2740726 | -0.0993491 |
| 102655896 | nucleoplasmin-like protein-like isoform X4 | 16.340267 | 0.1679860 | 0.1843370 | -0.1048845 | 0.0246945 |
| GB55474 | protein pygopus | 16.236962 | 0.2676167 | 0.0707588 | -0.0549138 | 0.0726924 |
| GB51125 | inositol-3-phosphate synthase 1-B isoform X2 | 16.234124 | -0.1324499 | 0.0450646 | -0.1206895 | -0.1431645 |
| GB45968 | collagen alpha-1(IV) chain-like isoform 1 | 16.142244 | -0.1021189 | -0.2851870 | -0.1437885 | -0.1852569 |
| GB44537 | myosin-IA | 16.130906 | -0.3302817 | -0.0513892 | 0.0332848 | -0.1087754 |
| GB45824 | phosphoserine phosphatase isoform X2 | 15.998310 | 0.1362108 | -0.0741826 | -0.1520385 | 0.9771061 |
| GB53567 | branched-chain-amino-acid aminotransferase, cytosolic-like isoform 1 | 15.898775 | -0.1946488 | -0.1506045 | -0.1192245 | 0.0252445 |
| 724293 | protein yellow | 15.838484 | 0.1487303 | -0.2653227 | 0.9050534 | 0.0119983 |
| GB44138 | l-2-hydroxyglutarate dehydrogenase, mitochondrial-like isoform X3 | 15.463787 | -0.0605374 | -0.0645523 | -0.0579366 | -0.0117976 |
| GB45975 | LIM/homeobox protein Lhx3 | 15.448320 | -1.4912892 | 0.1699440 | -0.5350890 | -0.2095720 |
| GB44420 | hydroxymethylglutaryl-CoA synthase 1 isoform X2 | 15.447539 | 0.0319813 | -1.4782209 | -0.2339946 | -0.2200166 |
| GB43942 | putative serine protease K12H4.7-like isoform X2 | 15.444385 | 0.6725080 | 0.0163704 | -0.1396986 | 0.1787550 |
| GB42629 | chromatin accessibility complex protein 1-like | 15.273897 | 0.1251692 | 0.1625803 | -0.0870455 | -0.0091544 |
| GB42541 | carbonic anhydrase-related protein 10-like isoform X3 | 15.217357 | -0.0902229 | -0.2070180 | -0.4223262 | -0.1162996 |
| GB54391 | putative glycogen [starch] synthase-like isoform X1 | 15.171482 | -0.1572320 | -0.1100485 | -0.0326887 | -0.1545854 |
| GB52496 | epoxide hydrolase 4-like isoform X4 | 15.110661 | 0.2774522 | 0.0115722 | -0.3747157 | -0.0800490 |
| GB51598 | translocon-associated protein subunit beta isoform 2 | 14.774053 | 0.5583941 | 0.0371216 | -0.0403127 | -0.1591828 |
| GB49095 | high affinity copper uptake protein 1-like isoformX1 | 14.620522 | 0.5230884 | -0.1010762 | -0.2729704 | -0.0231513 |
| GB54888 | 2-acylglycerol O-acyltransferase 1-like isoform X1 | 14.550168 | 0.6685157 | 0.0466928 | -0.0917668 | -0.1179655 |
| GB42264 | myb-like protein X-like | 14.521676 | -0.6252822 | -0.1368822 | 1.0730476 | -0.0070602 |
| GB45943 | collagen alpha-5(IV) chain | 14.385447 | -0.0141205 | -0.1686619 | -0.0306632 | 0.1661617 |
| GB51236 | acyl-CoA Delta(11) desaturase isoform X2 | 14.282561 | 0.7514364 | 0.0473029 | -0.6010006 | 0.4886544 |
| GB47503 | delta-1-pyrroline-5-carboxylate synthase-like isoform X3 | 14.246721 | -0.4533711 | -0.1181787 | -0.0922763 | -0.0916646 |
| GB47839 | calumenin | 14.227729 | 0.2508064 | 0.1073882 | 1.1338349 | 0.6543477 |
| GB40747 | GMP reductase 2-like isoform 1 | 14.074250 | -0.1389808 | -0.0848805 | 0.0539909 | 0.0060923 |
| GB55661 | neuronal membrane glycoprotein M6-a-like isoform X2 | 13.870018 | 0.2916694 | 0.0445663 | 0.0406151 | 0.0845796 |
| GB49854 | alpha-amylase precursor | 13.835878 | 0.4857671 | 0.0835219 | -1.6759631 | 0.9168179 |
| 726965 | uncharacterized protein LOC726965 | 13.828477 | -0.1511445 | -0.1317802 | 0.1458579 | -0.3221607 |
| 102655415 | uncharacterized protein LOC102655415 | 13.761128 | -0.1886490 | -0.2064529 | -0.8759375 | -0.3895903 |
| GB52114 | protein trachealess-like isoform X7 | 13.706700 | -0.6009081 | -0.0822315 | 2.8207841 | 0.0081549 |
| GB43216 | uncharacterized protein LOC413583 isoform X2 | 13.521796 | -0.2362268 | -0.1658183 | -0.4291904 | -0.6447770 |
| GB47449 | nucleoporin NUP188 homolog | 13.432553 | 0.0398209 | 0.1257927 | -0.3579522 | -0.0552367 |
| GB53230 | adipokinetic hormone receptor | 13.346403 | -0.1559642 | 0.0488804 | -0.4935836 | -0.3911266 |
| GB42738 | protein cueball-like | 13.218575 | 0.0792556 | 0.0187766 | -0.3277604 | -0.1859564 |
| GB42468 | phospholipase B1, membrane-associated-like isoform X1 | 13.019295 | 0.1712758 | -2.6550613 | -0.2391406 | 0.1282518 |
| GB48521 | RNA polymerase II elongation factor ELL2-like isoform X1 | 12.917540 | -0.0107311 | -0.1580317 | -0.1465078 | 0.0207912 |
| GB49321 | D-arabinitol dehydrogenase 1-like | 12.778844 | 0.3850348 | -0.0314272 | -0.1281534 | -0.1467144 |
| GB53404 | protein fork head-like isoform 1 | 12.776526 | -2.0510009 | 0.0551431 | 0.4763584 | -0.2108822 |
| 411557 | protein FAM46A-like isoformX2 | 12.640439 | -0.3724492 | 0.1541695 | -0.1698355 | -0.0530527 |
| GB44850 | origin recognition complex subunit 3-like | 12.544410 | 0.2060399 | -0.0628445 | -0.2511858 | 0.1533299 |
| GB51077 | dystrotelin-like isoform X1 | 12.364985 | -0.2613290 | 0.1545091 | -0.4736396 | 0.0442161 |
| GB49543 | alanine–glyoxylate aminotransferase 2-like | 12.351536 | 0.3435971 | -0.1281697 | -0.1746713 | -0.5539103 |
| GB52712 | serine/arginine repetitive matrix protein 2-like isoform X1 | 12.310149 | 0.1347869 | 0.2130550 | -0.1568323 | 0.1329923 |
| GB53036 | serine/threonine-protein kinase Warts-like isoform X1 | 12.294727 | -0.1758171 | 0.0278491 | -0.2505460 | 0.0098058 |
| GB46917 | uncharacterized protein LOC726071 | 12.249764 | 0.1372400 | 0.0917500 | -0.1651023 | 0.1279429 |
| GB53661 | methyltransferase-like isoform X3 | 12.132568 | -0.2274413 | -0.0795270 | 2.0586400 | -0.0170579 |
| GB51278 | innexin inx3 | 12.091606 | 0.3830713 | 0.0406630 | 0.0169686 | -0.1518675 |
| GB52161 | cuticular protein 28 precursor | 11.933221 | 0.2839393 | -0.6850552 | 0.0492736 | -0.0220819 |
| GB42887 | protein NPC2 homolog | 11.895962 | 0.4934256 | 0.1171569 | 0.8267574 | -0.2535760 |
| GB43984 | xenotropic and polytropic retrovirus receptor 1 homolog | 11.711923 | -0.0394515 | 0.0389923 | 0.0492826 | 0.1485743 |
| GB48252 | dihydrofolate reductase isoform X2 | 11.676320 | 0.2476259 | 0.2239091 | -0.1962494 | 0.1041595 |
| GB47270 | cytochrome P450 4C1 | 11.593087 | 0.3133208 | 0.1872647 | 0.7449695 | 0.5041494 |
| GB48109 | retinoid-inducible serine carboxypeptidase-like isoform X3 | 11.544591 | 0.9904184 | 0.1527256 | -0.4756137 | 0.2243044 |
| GB54313 | uncharacterized protein LOC413386 isoform X3 | 11.417055 | -0.2681125 | 0.0458400 | -0.5743109 | -0.0318130 |
| GB51913 | thymidylate kinase-like isoform X2 | 11.334845 | -0.3730554 | 0.3081842 | -0.1084622 | 0.0965259 |
| GB44503 | uncharacterized protein LOC727423 isoform X2 | 11.287468 | 0.8326731 | -0.2057741 | -0.1159152 | 0.0800218 |
| GB53229 | WAS protein family homolog 1-like | 11.254827 | 0.0009783 | 0.1385308 | 0.0844486 | -0.0924363 |
| GB51834 | sodium-dependent nutrient amino acid transporter 1-like | 11.166136 | -0.3531060 | -0.2620501 | 0.0808984 | 0.0511710 |
| GB52505 | chaoptin-like | 10.968009 | 0.4587636 | -0.0083482 | -0.3768354 | 0.1096745 |
| GB52275 | pancreatic lipase-related protein 2-like | 10.793842 | 0.1098269 | 0.2574445 | -0.4376261 | 0.1596069 |
| GB55302 | trehalose transporter 1 isoform X6 | 10.667963 | -0.0677498 | -0.1155692 | -0.2214355 | 0.3081366 |
| 102654789 | uncharacterized protein LOC102654789 | 10.667109 | -0.0584019 | -0.3278716 | -0.6095776 | 0.0498065 |
| GB42616 | beta-hexosaminidase subunit beta-like | 10.368794 | -0.2289562 | 0.1451877 | 0.0621008 | 0.0620511 |
| GB54153 | uncharacterized protein LOC100576236 isoform X1 | 10.118213 | -0.0809921 | -0.1347052 | -1.4023742 | -0.1316166 |
| GB47327 | lipid phosphate phosphohydrolase 3-like | 10.058571 | 0.2857168 | -0.2176489 | 0.1057685 | -0.0934929 |
| GB50021 | exonuclease 3’-5’ domain-containing protein 2-like isoform X1 | 9.842047 | -0.0736605 | 0.2350361 | 0.2707233 | 0.0938046 |
| GB40344 | uncharacterized protein LOC552242 | 9.828621 | 0.1172944 | -0.8017426 | -0.2242850 | 0.5032930 |
| GB49929 | laminin subunit alpha | 9.561491 | 0.5340516 | 0.2430509 | -0.5370697 | -0.6102948 |
| GB44663 | homeobox protein Nkx-2.4-like | 9.551657 | 0.1394744 | -0.3654465 | 0.1719647 | -0.1904368 |
| GB51107 | uncharacterized protein LOC100578731 isoform X1 | 9.377499 | 0.4302073 | 0.1834154 | 0.1378654 | -0.1713280 |
| GB50524 | uncharacterized protein LOC726417 | 8.929495 | -0.0588231 | 0.0511584 | -1.3909039 | -0.1613334 |
| GB51696 | hexamerin 70c precursor | 8.783477 | -0.0474407 | -0.2625302 | 0.0807997 | 0.5193354 |
| GB51195 | protein abrupt-like isoform X5 | 8.448029 | -0.0648071 | 0.2780828 | -0.3448047 | 0.0069477 |
| GB46800 | uncharacterized protein LOC100577231 | 8.330772 | -0.6305159 | -0.0697162 | 0.5306095 | 0.0739981 |
| GB52656 | uncharacterized protein LOC552154 | 8.299683 | 0.5834384 | -0.2149017 | -0.5531551 | -0.1221217 |
| GB42799 | protein takeout-like | 8.250769 | -0.2869070 | 0.2517661 | -1.5197911 | -1.0284651 |
| GB42426 | glutamyl aminopeptidase-like isoform X2 | 7.939937 | 0.7268796 | 0.4365634 | -0.6505219 | 0.0556743 |
| GB53155 | maternal embryonic leucine zipper kinase-like | 7.859620 | -0.0641905 | 0.0686990 | 3.1684473 | 0.0911321 |
| GB44967 | GTP:AMP phosphotransferase AK3, mitochondrial isoform X1 | 7.715168 | -0.2241576 | -0.0260340 | 1.0680177 | 0.3088382 |
| GB48079 | trypsin-7 | 7.662906 | 0.7889055 | -0.6491093 | -0.9266060 | 0.0966946 |
| 102656088 | uncharacterized protein LOC102656088 | 7.565171 | 1.6565192 | -0.7683197 | -0.7692478 | -0.7298828 |
| GB50434 | proton-coupled amino acid transporter 1-like | 7.268236 | 0.0112121 | -0.1261022 | -0.0116264 | -0.2000854 |
| GB49813 | SUMO-activating enzyme subunit 1 | 7.001295 | 0.5203949 | 0.2339421 | 0.6507883 | 0.1731960 |
| GB43181 | uncharacterized protein LOC552799 isoform X2 | 6.971888 | 0.5939354 | 0.7192746 | -0.8612463 | 0.0416577 |
| GB46693 | WD repeat-containing protein 65-like | 6.867550 | 0.1122710 | -0.0684919 | 0.1286625 | -0.1355103 |
| GB47181 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | 6.863152 | -0.1128277 | -0.0754332 | 0.5035167 | 0.1284447 |
| GB52667 | uncharacterized protein LOC552202 isoform X6 | 6.664971 | -0.2443954 | -0.0018447 | -1.1119964 | -0.0773408 |
| GB53401 | protein fosB isoform X1 | 6.443355 | 0.0228858 | -0.1389772 | 0.0142738 | 0.1133762 |
| GB47507 | histone H2A-like | 6.019835 | -0.6332116 | 0.2794373 | -0.2087029 | -0.1453361 |
| GB45458 | UDP-glucose 6-dehydrogenase-like isoform X2 | 5.705610 | -0.0389463 | -0.0525780 | -0.3343070 | -0.2095720 |
| GB41782 | LOW QUALITY PROTEIN: glycine dehydrogenase [decarboxylating], mitochondrial-like | 5.448141 | 0.1849845 | -0.2525962 | -0.0070789 | -0.1010127 |
| GB54426 | transmembrane protein 205-like | 5.184689 | -0.0505523 | 0.1720915 | 0.0864230 | -0.0824220 |
| GB43591 | uncharacterized protein LOC408443 | 4.927475 | 0.4363105 | 0.2200117 | -0.3238923 | 0.1074204 |
| GB46298 | endocuticle structural glycoprotein SgAbd-8-like isoform X2 | 3.339703 | 0.2037923 | -0.6881852 | -0.0825740 | 0.1612789 |
Module 5
Table S30: List of all the genes in Module 5, ranked by their within-module connectivity, k. The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
gene_list <- module_gene_list(5)
saveRDS(gene_list, file = "supplement/tab_S30.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB51221 | ubiquitin-protein ligase E3A isoform X2 | 24.892744 | 0.4052710 | 0.0577656 | 0.0239629 | 0.0567963 |
| GB53171 | transcription elongation factor B polypeptide 2 | 23.618178 | 0.1968781 | 0.0123665 | -0.0062142 | 0.0805982 |
| GB55522 | RING finger protein 11-like isoform X2 | 21.611563 | -0.0310051 | 0.0292602 | 0.0694682 | 0.0424390 |
| GB50246 | membrane-associated protein Hem | 20.447362 | 0.2836203 | -0.0255881 | 0.0093727 | 0.1126247 |
| GB55382 | putative oxidoreductase GLYR1 homolog | 20.162157 | -0.0285636 | -0.0377781 | -0.0158068 | -0.0035803 |
| GB50020 | signal transducer and activator of transcription 5B | 19.693792 | 0.1536933 | -0.0559117 | -0.0076774 | 0.0551949 |
| GB47341 | protein disulfide-isomerase TMX3-like isoform X2 | 19.376720 | 0.2339978 | 0.0667786 | -0.0142374 | 0.1435044 |
| GB54119 | vacuolar protein sorting-associated protein 11 homolog isoform X1 | 19.075278 | 0.3001633 | -0.0495528 | 0.0161061 | 0.0935256 |
| GB52529 | F-actin-capping protein subunit beta-like | 18.873344 | 0.3099505 | -0.0217876 | 0.0087232 | 0.0522579 |
| GB44934 | nuclear cap-binding protein subunit 1 isoformX1 | 18.351405 | 0.0571397 | 0.0622121 | -0.0055183 | 0.1911069 |
| GB41553 | Golgi phosphoprotein 3 homolog rotini-like isoform X1 | 18.293273 | 0.4552440 | -0.0487894 | 0.0548868 | 0.0138926 |
| GB44725 | uncharacterized protein LOC552067 | 18.170921 | 0.7334216 | -0.0241969 | -0.0485013 | 0.1149952 |
| GB52497 | mitochondrial import receptor subunit TOM20 homolog | 18.003825 | 0.2545658 | -0.0054186 | -0.0495156 | 0.0974993 |
| GB43692 | CCR4-NOT transcription complex subunit 11-like | 17.935755 | 0.2380759 | -0.0868604 | -0.0228156 | 0.1579734 |
| GB50194 | nuclear inhibitor of protein phosphatase 1 isoform X2 | 17.724071 | 0.0891565 | 0.1432309 | 0.0276218 | 0.0529888 |
| GB54129 | protein LSM12 homolog A-like isoform X2 | 17.630725 | 0.1263633 | 0.0047431 | 0.0084219 | 0.0954643 |
| GB43194 | syntaxin-8 isoform 2 | 17.620125 | 0.0812011 | 0.0439155 | -0.0010292 | 0.1494998 |
| GB49114 | nucleolar complex protein 4 homolog B-like isoform X1 | 17.502510 | 0.4090210 | 0.0106571 | 0.0459992 | 0.1265175 |
| GB45042 | neuroguidin-A-like | 17.294846 | -0.0865711 | -0.0133114 | 0.0818932 | 0.1217293 |
| GB44445 | 5’-3’ exoribonuclease 2 homolog | 17.285802 | 0.1452856 | 0.0030913 | -0.0506493 | 0.1907890 |
| GB55624 | leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 2-like isoform X2 | 17.131354 | 0.1718467 | 0.0943323 | 0.0381437 | 0.0596429 |
| GB42559 | oxysterol-binding protein 1 isoform X5 | 17.015087 | 0.2195971 | 0.0247663 | 0.0291211 | -0.1329023 |
| GB45368 | serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform isoform 1 | 16.895902 | 0.2668460 | 0.0491584 | 0.0465789 | 0.1523685 |
| GB44299 | TOM1-like protein 2-like isoform X3 | 16.832881 | 0.0597279 | -0.0063979 | 0.0377341 | 0.0161815 |
| GB53245 | UBX domain-containing protein 6 | 16.806675 | 0.1492805 | -0.0494149 | -0.0321533 | 0.1686399 |
| GB41069 | pyridoxal-dependent decarboxylase domain-containing protein 1-like isoform X2 | 16.772074 | 0.5433672 | -0.0623971 | -0.0427447 | 0.0339210 |
| GB53770 | 39S ribosomal protein L51, mitochondrial | 16.717284 | 0.0713392 | -0.0122901 | 0.0315855 | 0.1599665 |
| GB54381 | lisH domain-containing protein C1711.05-like isoform X1 | 16.624306 | 0.0356870 | 0.0812525 | 0.6978276 | 0.0568396 |
| GB44580 | nucleolysin TIAR | 16.427377 | 0.1807535 | 0.0398625 | -0.0181675 | 0.6162550 |
| GB42961 | glutathione S-transferase theta-4 | 16.398054 | 0.2744631 | -0.1522298 | -0.0996587 | 0.1190441 |
| GB44067 | probable phospholipid-transporting ATPase IIB-like | 16.330738 | 0.0810361 | -0.0094281 | 0.0266179 | 0.2117633 |
| GB46465 | alpha/beta hydrolase domain-containing protein 13-like isoform X1 | 16.287041 | 0.0431816 | 0.1021687 | -0.0729202 | 0.1067043 |
| GB52655 | signal transducing adapter molecule 1 | 16.013912 | 0.0488033 | 0.0109904 | -0.0075249 | 0.0406170 |
| GB50471 | cation-independent mannose-6-phosphate receptor isoform X2 | 15.866288 | 0.1863032 | -0.1386914 | -0.0873615 | 0.0281946 |
| GB49399 | COP9 signalosome complex subunit 4 isoform X2 | 15.854596 | 0.1634535 | 0.1089935 | -0.0585791 | 0.1899583 |
| GB46766 | F-actin-capping protein subunit alpha-like | 15.850821 | 0.2751667 | 0.0220082 | -0.0215659 | 0.1275786 |
| GB51186 | probable Ufm1-specific protease 2-like isoform X1 | 15.840711 | 0.1177674 | -0.0440234 | 0.0752169 | 0.1297003 |
| GB50701 | pleckstrin homology domain-containing family M member 1-like | 15.645477 | 0.0932321 | -0.1190251 | -0.3553708 | 0.0360539 |
| GB40598 | LOW QUALITY PROTEIN: bumetanide-sensitive sodium-(potassium)-chloride cotransporter | 15.493503 | 0.0513337 | 0.0614736 | 0.0163903 | 0.0251189 |
| GB41688 | cullin-1-like isoformX1 | 15.485842 | 0.1166784 | 0.1107078 | 0.0292815 | 0.1933199 |
| GB40451 | alanine aminotransferase 2-like | 15.330310 | 0.2675747 | -0.0980800 | -0.0813782 | 0.1356313 |
| GB43438 | WD repeat-containing protein 89-like | 15.321326 | 0.2380182 | 0.1053031 | -0.0396475 | 0.1473739 |
| GB55444 | serine/threonine-protein phosphatase PP1-beta-like isoform 1 | 15.310679 | 0.1203893 | 0.0398922 | -0.0403059 | 0.1171705 |
| GB50802 | putative tyrosine-protein kinase Wsck-like | 15.306046 | -0.1967245 | -0.0351351 | 0.0168308 | 0.0036224 |
| GB55431 | ubiquitin-conjugating enzyme E2 R2-like isoform X1 | 15.131201 | 0.2234337 | 0.1204319 | -0.0951763 | 0.1041750 |
| GB42155 | craniofacial development protein 1 isoform X1 | 15.106298 | 0.1132166 | 0.0889102 | -0.0046777 | 0.2277060 |
| GB41063 | protein ariadne-1-like isoform X1 | 15.035359 | -0.0939936 | -0.0900601 | 0.0227550 | 0.1635534 |
| GB45350 | zinc finger CCCH domain-containing protein 10-like isoform X1 | 15.001063 | 0.1088458 | 0.0497915 | 0.1276458 | 0.1081093 |
| GB44037 | ATP-dependent RNA helicase Ddx1 isoform 1 | 14.829007 | 0.2734065 | 0.0444311 | 0.1005937 | -0.2979170 |
| GB51197 | protein TSSC1-like | 14.734155 | 0.2349185 | -0.1430111 | -0.0833998 | 0.2667779 |
| GB42241 | membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 isoform X6 | 14.711949 | 0.3906202 | -0.0426757 | -0.0171553 | 0.0893601 |
| GB49353 | lamin Dm0-like isoform X2 | 14.657339 | 0.1673704 | 0.0545500 | 0.3564577 | -0.0715816 |
| GB44496 | probable serine incorporator isoformX1 | 14.488674 | 0.3760626 | -0.0164596 | 0.0240435 | 0.1268478 |
| GB40807 | uncharacterized protein LOC724585 | 14.462108 | 0.3215229 | 0.0310074 | -0.1273385 | 0.1049413 |
| 102655399 | proteasome assembly chaperone 4-like | 14.418063 | 0.3303956 | 0.0649544 | 0.0421997 | 0.2416008 |
| GB48926 | NEDD8-conjugating enzyme Ubc12-like | 14.366897 | 0.4077934 | 0.0745049 | 0.0494170 | 0.1154497 |
| GB40888 | CAAX prenyl protease 1 homolog isoform X3 | 14.233110 | 0.3782520 | -0.0698724 | -0.0556332 | 0.0337629 |
| 102656934 | stress-induced-phosphoprotein 1-like | 13.986812 | 0.2938157 | 0.1163782 | -0.3097045 | 0.2531416 |
| GB40727 | CDP-diacylglycerol–inositol 3-phosphatidyltransferase-like | 13.966864 | 0.3473500 | -0.1168118 | 0.0362526 | 0.1023306 |
| GB43905 | stomatin-like protein 2, mitochondrial-like isoform 1 | 13.955950 | 0.2195851 | 0.0976619 | 0.0626404 | 0.1568970 |
| GB48404 | transmembrane and coiled-coil domains protein 1-like isoform X1 | 13.925877 | 0.1440461 | 0.1527323 | -0.0775174 | 0.2831018 |
| GB42952 | ADP-ribosylation factor-like protein 1-like isoform 1 | 13.875415 | 0.4526867 | -0.0721123 | -0.0340103 | 0.1755959 |
| GB43554 | adapter molecule Crk-like isoform X2 | 13.845748 | 0.1506577 | 0.0182856 | -0.0174315 | 0.0498358 |
| GB55295 | bis(5’-nucleosyl)-tetraphosphatase [asymmetrical] isoform X4 | 13.767365 | 0.5171171 | -0.0299657 | -0.0571674 | 0.1346402 |
| GB53438 | histone deacetylase Rpd3 isoform 1 | 13.656368 | 0.1847471 | 0.0490797 | -0.1190312 | 0.2379871 |
| GB52745 | thioredoxin domain-containing protein 15-like isoform X3 | 13.598898 | 0.1682122 | 0.0887100 | -0.0268504 | 0.1118756 |
| GB47463 | guanine nucleotide-binding protein G(i) subunit alpha-like | 13.577702 | 0.5054963 | 0.0186759 | -0.0877653 | 0.1429009 |
| GB50980 | acyl-protein thioesterase 1-like | 13.541260 | 0.5521331 | -0.0028859 | 0.0556054 | 0.1087179 |
| GB55852 | E3 ubiquitin-protein ligase RNF185-like isoform 1 | 13.518647 | 0.2727766 | -0.1636059 | -0.0425730 | 0.0836839 |
| GB44896 | coiled-coil domain-containing protein 132-like | 13.403985 | 0.3915606 | -0.0158847 | -0.0855921 | 0.1037450 |
| GB55090 | probable ATP-dependent RNA helicase DDX56 isoform 1 | 13.300122 | 0.4103054 | 0.0706519 | -0.0638143 | 0.2355292 |
| GB40309 | peroxisomal biogenesis factor 19-like isoform X1 | 13.238939 | -0.0434331 | -0.1190064 | 0.0166376 | 0.1790234 |
| GB40895 | WD repeat domain-containing protein 83-like isoform 1 | 13.237225 | 0.2042820 | 0.0477285 | -0.0867027 | 0.1374369 |
| GB53380 | uncharacterized protein KIAA2013 homolog | 13.118861 | 0.2251021 | -0.0726437 | -0.0273242 | 0.1061084 |
| GB42694 | AP-1 complex subunit mu-1-like isoform 1 | 13.030828 | 0.2547400 | 0.0937519 | 0.1195458 | 0.0275934 |
| GB43203 | niemann-Pick C1 protein-like isoform X3 | 12.973766 | 0.3944177 | 0.0017272 | -0.0639502 | 0.0348854 |
| 724366 | ras-related protein Rap-2a | 12.910182 | 0.0892249 | -0.0176146 | 0.0108156 | 0.6263025 |
| GB42845 | adenylate kinase | 12.907800 | 0.1358788 | -0.1425863 | -0.0668998 | 0.0489313 |
| GB42266 | UPF0472 protein C16orf72 homolog | 12.906588 | 0.0437115 | -0.1873511 | -0.1043398 | 0.0963684 |
| GB52191 | mannan-binding lectin serine protease 1 | 12.893436 | 0.2440224 | 0.0861990 | -0.2521358 | 0.0069322 |
| GB42944 | ras-related protein Rab-10 isoformX2 | 12.806989 | 0.2822878 | 0.0571568 | -0.0433214 | 0.2128106 |
| GB40289 | ATP-dependent (S)-NAD(P)H-hydrate dehydratase-like isoform X1 | 12.785933 | 0.4984351 | 0.0064125 | -0.2263815 | 0.0836054 |
| GB50075 | probable palmitoyltransferase ZDHHC16-like isoform X1 | 12.762470 | -0.1051041 | 0.1535196 | -0.0512994 | 0.2251726 |
| GB41035 | ubiquitin carboxyl-terminal hydrolase 31-like isoform 1 | 12.685733 | -0.2197074 | 0.1882218 | -0.0399316 | 0.1448446 |
| GB42022 | ubiquitin carboxyl-terminal hydrolase 34-like | 12.645357 | 0.1043158 | -0.1179329 | 1.0013935 | 0.0335301 |
| GB43184 | iron/zinc purple acid phosphatase-like protein-like | 12.628967 | 0.4506253 | 0.0972285 | 0.4273176 | 0.1708128 |
| GB45944 | ras-related protein Rab-14 isoform 1 | 12.588465 | 0.0657949 | -0.0256554 | -0.0103594 | 0.1635179 |
| GB52762 | ceramide-1-phosphate transfer protein-like isoform X3 | 12.377700 | 0.4564015 | 0.1179392 | 0.1449446 | 0.1999809 |
| GB42416 | solute carrier family 35 member C2-like isoform X3 | 12.367298 | 0.4198498 | 0.1962084 | -0.0589713 | 0.0170164 |
| GB46768 | uncharacterized MFS-type transporter C09D4.1-like isoform X6 | 12.338673 | 0.1790280 | 0.0055079 | -0.0632106 | 0.0522802 |
| GB42940 | protein lifeguard 1-like isoform X3 | 12.324248 | 0.2462224 | -0.1035382 | 0.4238047 | 0.0547987 |
| GB41046 | ubiquitin-conjugating enzyme E2 Q2-like isoform X1 | 12.309335 | 0.1483677 | 0.0664233 | -0.0398509 | 0.0738568 |
| GB49529 | proliferation-associated protein 2G4-like | 12.301328 | 0.5851265 | 0.0735879 | -0.0864866 | -0.4536047 |
| GB40339 | iodotyrosine dehalogenase 1-like isoformX2 | 12.298492 | 0.2152316 | 0.2089693 | -0.0579476 | 0.0308205 |
| GB55478 | probable E3 ubiquitin-protein ligase makorin-1 isoform X2 | 12.284477 | 0.0474732 | 0.1391572 | 0.0093410 | 0.1430543 |
| GB54251 | DCN1-like protein 1-like isoform X2 | 12.271228 | -0.1746568 | -0.0525520 | -0.0614046 | 0.0812937 |
| GB50219 | DE-cadherin-like isoform X5 | 12.184665 | 0.1788053 | 0.0892066 | -0.7342080 | -0.4583919 |
| GB43103 | transmembrane emp24 domain-containing protein 5 | 11.984311 | 0.5031756 | 0.1400732 | 0.0115312 | -0.2290611 |
| GB45558 | ras-related protein Rac1 isoform 1 | 11.982039 | 0.1710494 | -0.0665692 | -0.0154981 | -0.0008272 |
| GB51358 | TIP41-like protein-like isoform X2 | 11.943016 | 0.2155564 | 0.2026226 | 0.0126609 | 0.1848117 |
| GB52754 | probable dimethyladenosine transferase-like | 11.813125 | 0.3191406 | 0.0929833 | -0.1781242 | 0.1720469 |
| 102654186 | fizzy-related protein homolog | 11.796964 | 0.4751686 | -0.1062848 | -0.0719079 | 0.0622565 |
| GB44869 | soluble NSF attachment protein isoform X2 | 11.738834 | 0.4330352 | -0.1460116 | 0.0372161 | 0.1644114 |
| GB44773 | neural Wiskott-Aldrich syndrome protein isoform X3 | 11.683791 | 0.4673480 | 0.1287391 | -0.1127766 | 0.1158484 |
| GB44181 | E3 ubiquitin-protein ligase RNF8-like isoform X2 | 11.682968 | 0.6945363 | 0.0524556 | 0.0049124 | 0.3925765 |
| GB49107 | uncharacterized threonine-rich GPI-anchored glycoprotein PJ4664.02 | 11.682337 | 0.0065103 | 0.0214853 | -0.0278378 | 0.1423447 |
| GB55581 | membrane-associated progesterone receptor component 1-like isoform 2 | 11.634351 | 0.5127324 | -0.1137708 | -0.0437872 | -0.2402691 |
| GB54789 | GMP synthase [glutamine-hydrolyzing] | 11.614943 | 0.3706741 | 0.0244532 | -0.0489558 | 0.0349607 |
| GB50475 | E3 ubiquitin-protein ligase RAD18-like | 11.541742 | 0.1463569 | 0.1750133 | -0.0056663 | 0.3141866 |
| GB47753 | transmembrane protein 208-like | 11.527285 | 0.1838047 | 0.0967788 | -0.0306899 | -0.0837817 |
| GB53819 | uncharacterized protein LOC100577293 | 11.514756 | 0.0483880 | 0.1343646 | 1.6786168 | 0.2222360 |
| GB46586 | protein phosphatase 1H-like | 11.464911 | 0.0706660 | -0.0408080 | 1.4936484 | 0.2889797 |
| GB43135 | RAC serine/threonine-protein kinase | 11.442577 | 0.2443803 | -0.0003408 | 0.0821850 | 0.1752096 |
| GB46333 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1-like | 11.434899 | 0.2910376 | 0.1127519 | 0.0426768 | 0.1299636 |
| GB40911 | E3 SUMO-protein ligase PIAS3 isoform X2 | 11.429816 | 0.0966192 | 0.1167597 | -0.0272543 | 0.1145917 |
| GB52059 | eukaryotic translation initiation factor 4H-like isoform X1 | 11.423621 | 0.3708224 | 0.0556411 | 4.7089102 | 0.0874739 |
| GB46567 | eukaryotic translation initiation factor 1A, X-chromosomal | 11.355016 | 0.3296399 | -0.0054643 | -0.3233193 | 0.3985244 |
| GB53043 | ATP-binding cassette sub-family G member 4 isoform X2 | 11.317094 | 0.1882006 | -0.1872264 | 0.1764359 | 0.1029270 |
| GB43843 | thioredoxin-related transmembrane protein 2 homolog | 11.207869 | -0.0542081 | 0.1875795 | -0.2917607 | 0.2665958 |
| GB49351 | solute carrier family 35 member E2-like isoform X1 | 11.156500 | 0.0484972 | 0.0724831 | 0.1008705 | 0.1497114 |
| GB40126 | uncharacterized protein LOC410456 isoform X1 | 11.136334 | -0.3292055 | -0.1095215 | -0.1067866 | 0.2848831 |
| GB53284 | proto-oncogene tyrosine-protein kinase receptor Ret-like isoform X3 | 11.118872 | 0.5298880 | -0.0293171 | -0.1119531 | -0.4666892 |
| GB42488 | vesicle transport protein GOT1B-like isoform 2 | 11.101933 | 0.5833012 | 0.0050660 | -0.1791475 | 0.0852511 |
| GB54950 | uncharacterized protein LOC726431 isoform X1 | 11.099108 | -0.0943708 | -0.1289242 | -1.0586067 | 0.1886479 |
| GB42797 | protein takeout-like | 11.034881 | 1.3020633 | 0.1495762 | 0.4395761 | 0.1539929 |
| GB47542 | eukaryotic translation initiation factor 3 subunit J isoform 1 | 10.986213 | 0.3985097 | -0.0161926 | -0.1284190 | 0.1974344 |
| GB45071 | G-protein-signaling modulator 2 | 10.924741 | 0.2983616 | -0.1176442 | 0.0697734 | 0.1776567 |
| GB54449 | DDB1- and CUL4-associated factor 7-like | 10.845327 | 0.5835080 | -0.1107505 | 0.0575219 | 0.2315420 |
| GB42106 | probable histone-binding protein Caf1 | 10.820084 | 0.4923891 | 0.2027339 | -0.1112418 | 0.1288082 |
| GB41158 | toll-interacting protein | 10.818064 | 0.5070158 | -0.0545298 | 0.2897440 | 0.1457877 |
| GB44621 | serine/threonine-protein phosphatase 5 | 10.702357 | -0.0169020 | -0.0501653 | -0.9193905 | 0.0935470 |
| GB42084 | probable G-protein coupled receptor Mth-like 1-like isoform X2 | 10.547147 | 0.5477811 | -0.0984398 | 0.0638015 | -0.0491167 |
| GB53627 | zinc transporter ZIP9-B-like | 10.490326 | 0.6344679 | -0.0820151 | -0.0146221 | 0.1952920 |
| GB42313 | leishmanolysin-like peptidase | 10.383085 | 0.4577268 | -0.0837123 | -0.0803312 | 0.3368716 |
| GB47391 | nuclear pore complex protein Nup107 | 10.264407 | -0.0721120 | 0.1623645 | -0.0978919 | 0.1421264 |
| GB40708 | tetraspanin 6 isoform X1 | 10.221502 | 0.5640446 | -0.0485612 | 0.5170397 | 0.2038826 |
| GB44383 | histone acetyltransferase Tip60 | 10.070100 | 0.0591542 | 0.1431674 | -0.0248541 | 0.3156142 |
| GB41467 | growth arrest-specific protein 2-like isoform X2 | 10.055256 | 0.0062887 | -0.1447545 | -0.1951977 | 0.1293803 |
| GB51551 | myophilin | 9.649953 | 0.4369511 | 0.1042199 | -0.0373071 | 0.1759337 |
| GB48646 | ADP-ribosylation factor-like protein 2-like isoform 1 | 9.428523 | 0.3889522 | -0.0730585 | -0.0575960 | 0.1530031 |
| GB54084 | histone H3.3-like isoform 2 | 9.327566 | 0.5602693 | 0.1727171 | -0.2618429 | 0.1448937 |
| GB47231 | exportin-5 | 9.203596 | 0.2447790 | 0.1130894 | -0.3281473 | 0.0297004 |
| GB54172 | sodium-independent sulfate anion transporter-like isoformX1 | 9.122333 | 0.6638947 | 0.0879634 | -0.0240818 | 0.0611113 |
| GB45313 | uncharacterized protein LOC552058 isoform X4 | 9.063337 | -0.0071691 | -0.3387229 | 0.0326936 | 0.2449850 |
| GB42817 | transmembrane protein 63B-like isoform X3 | 9.063263 | 0.3831193 | 0.0266120 | 0.0829623 | 0.0291628 |
| GB42168 | receptor expression-enhancing protein 5-like isoform X2 | 8.991124 | 0.6230287 | -0.0920617 | -0.3059799 | -0.4304387 |
| GB55219 | uncharacterized protein LOC724286 isoform X2 | 8.563900 | 0.4319688 | -0.1544041 | 0.0813073 | 0.1107950 |
| GB54052 | 72 kDa inositol polyphosphate 5-phosphatase-like isoform X1 | 8.323068 | 0.0388923 | -0.0213879 | 0.6643017 | -0.1477970 |
| GB40758 | icarapin-like | 7.941814 | 0.4280829 | 0.0700838 | -0.2444572 | 0.1076566 |
| GB41793 | cytochrome c-type heme lyase-like | 7.877639 | 0.0394344 | -0.1229161 | 0.0258532 | 0.2649210 |
| GB43617 | uncharacterized membrane protein DDB_G0293934-like isoform X1 | 7.441465 | 0.6585730 | -0.0017682 | -0.0043774 | 0.7339717 |
| GB46795 | papilin-like isoform X7 | 7.089716 | 0.2051422 | -0.0964166 | 0.1142438 | 0.0292950 |
| 409791 | cAMP-dependent protein kinase catalytic subunit isoform 1 | 6.923792 | 0.1555292 | -0.2799863 | -0.0383467 | 0.1610000 |
| GB40838 | endoglucanase 15-like | 6.911462 | 0.3971706 | -0.1095881 | 0.3258507 | 0.2033069 |
| GB45983 | venom acid phosphatase Acph-1-like isoform X3 | 6.409778 | 0.4032656 | -0.0224067 | -0.0960330 | -0.0152726 |
| GB44192 | leucine-rich repeat-containing protein 26-like | 6.023099 | 0.4873011 | -0.3811280 | -0.1090735 | 0.1177434 |
| GB45973 | aromatic-L-amino-acid decarboxylase | 5.798389 | -0.0572646 | -0.2786491 | 0.0371271 | 0.1741154 |
| GB45673 | alpha-N-acetylgalactosaminidase-like | 4.999660 | 0.6834262 | 0.0267121 | 0.0716312 | 0.2765861 |
| GB42272 | Usher syndrome type-1G protein homolog isoform X3 | 4.748456 | 0.4716313 | 0.0090616 | 0.0329818 | 0.2845156 |
| GB40566 | cuticular protein 6 precursor | 4.625520 | 0.9719681 | -0.0805438 | -0.4304527 | 0.2944209 |
Module 6
Table S31: List of all the genes in Module 6, ranked by their within-module connectivity, k. The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
gene_list <- module_gene_list(6)
saveRDS(gene_list, file = "supplement/tab_S31.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB55009 | another transcription unit protein | 19.371985 | -0.0098566 | 0.1158533 | 0.0012225 | 0.1680409 |
| GB46605 | E3 ubiquitin-protein ligase CHIP-like isoform X2 | 19.333364 | -0.0610081 | 0.0838033 | 0.0520793 | 0.1626000 |
| GB42203 | putative adenosylhomocysteinase 3-like isoform X1 | 17.465953 | -0.1119988 | 0.0933100 | 0.0749205 | 0.1414484 |
| GB49412 | transcription factor IIIB 90 kDa subunit-like isoformX2 | 16.941039 | -0.2990613 | 0.0517704 | 0.1135770 | 0.1724694 |
| GB19050 | glutathione S-transferase C-terminal domain-containing protein | 16.582466 | -0.0738318 | 0.0695639 | 0.0640194 | 0.2401097 |
| GB52651 | diphthine–ammonia ligase-like isoform X4 | 16.257843 | -0.0199677 | 0.1052417 | 0.0638135 | 0.1738751 |
| GB54650 | protein PFC0760c-like isoform X1 | 16.231656 | -0.1221606 | -0.1833153 | 0.0457648 | 0.1728356 |
| GB44792 | PX domain-containing protein kinase-like protein-like isoform X3 | 15.895224 | -0.1449050 | -0.0745539 | 0.1138628 | 0.2064901 |
| GB41134 | Golgi SNAP receptor complex member 1 isoform 1 | 15.894398 | 0.0579986 | 0.1366296 | 0.0300338 | 0.1698680 |
| GB44828 | uncharacterized protein LOC550822 | 15.166371 | -0.0174765 | -0.1106948 | 0.0451509 | 0.2318739 |
| GB55940 | gamma-tubulin complex component 3-like | 15.160130 | -0.1656803 | -0.0064165 | 1.0465724 | 0.1705082 |
| GB47667 | dnaJ homolog subfamily C member 16-like | 15.071671 | -0.1804054 | 0.0528603 | 0.0701196 | 0.2845086 |
| GB49528 | exocyst complex component 6B isoform 1 | 14.846515 | -0.1644396 | 0.0498492 | 0.1198864 | 0.2661608 |
| GB52340 | uncharacterized protein LOC409246 | 14.741214 | -0.2853908 | -0.0047900 | 0.0363634 | 0.1110859 |
| GB42246 | rho GTPase-activating protein 190 isoform X3 | 14.583683 | -0.2032154 | 0.0418262 | 0.2870322 | 0.1626302 |
| 725733 | uncharacterized protein LOC725733 isoform X3 | 14.437612 | 0.0217862 | -0.0045017 | 0.0879935 | 0.3010459 |
| GB47841 | dnaJ homolog dnj-5-like | 14.271904 | -0.1231997 | 0.0602952 | 0.0851171 | 0.1738680 |
| GB44615 | extended synaptotagmin-1 isoform X1 | 14.246661 | 0.1117549 | -0.0251625 | 0.0934695 | -0.0473800 |
| GB46907 | zinc finger CCHC domain-containing protein 8 homolog | 14.234391 | -0.1888087 | 0.0353786 | 0.2435233 | 0.1590564 |
| GB40574 | serine palmitoyltransferase 1 | 13.720678 | -0.3901711 | -0.0928964 | 0.0157667 | 0.1601411 |
| GB42753 | ras-related protein Rab-7a-like | 13.633778 | 0.2007864 | -0.0655285 | 0.0926295 | 0.1155355 |
| GB41692 | DIS3-like exonuclease 2-like isoform X1 | 13.601457 | -0.1938238 | 0.1117541 | 0.0723061 | -1.0257958 |
| GB41292 | metallophosphoesterase 1 homolog, transcript variant X2 | 13.584422 | -0.3815023 | 0.0495884 | 0.1268113 | 0.1877302 |
| GB49413 | COP9 signalosome complex subunit 5 isoform X4 | 13.558844 | 0.1622229 | 0.0055072 | 0.0300382 | 0.1296849 |
| GB41738 | uncharacterized protein LOC410546 isoform X1 | 13.211043 | -0.1393708 | 0.2535065 | 0.1195139 | 0.1297026 |
| GB45296 | amyloid protein-binding protein 2 | 12.959462 | -0.0691975 | 0.0454775 | -0.0231756 | -0.0201450 |
| GB47837 | putative high mobility group protein 1-like 10-like isoform X1 | 12.942688 | 0.0485001 | -0.0301532 | 0.1609715 | 0.2440611 |
| GB50816 | ankyrin repeat domain-containing protein 54-like | 12.746179 | -0.1812111 | 0.1669438 | -0.1562977 | 0.0843522 |
| GB42375 | solute carrier family 25 member 46-like isoform 1 | 12.713325 | 0.1019241 | -0.0612529 | 0.0876314 | 0.1934626 |
| GB53222 | ER membrane protein complex subunit 7-like | 12.674148 | -0.0143630 | -0.0541264 | 0.0280253 | 0.1464846 |
| GB54588 | transport and Golgi organization protein 11 | 12.578910 | -0.0733121 | 0.0811735 | 0.0211903 | 0.1812808 |
| GB54210 | lipoma-preferred partner homolog isoform X4 | 12.554038 | -0.0022237 | 0.1214728 | 0.1480517 | 0.1090647 |
| GB44880 | DENN domain-containing protein 1A-like isoformX1 | 12.490344 | -0.0790916 | 0.0678043 | 0.4021882 | 0.1472870 |
| 102653759 | zinc finger CCHC domain-containing protein 4-like | 12.272363 | -0.5198039 | 0.1448227 | -0.1832482 | 0.3508098 |
| GB49366 | RNA polymerase II-associated protein 1-like | 12.231345 | -0.1111532 | 0.1335927 | 0.0160027 | 0.1019747 |
| GB54971 | BTB/POZ domain-containing protein 9 isoform X1 | 12.149088 | 0.0126651 | -0.0349103 | 0.1088447 | 0.1352351 |
| 412247 | putative ribosomal RNA methyltransferase CG11447-like | 12.126600 | -0.0682681 | 0.0401694 | -0.0020290 | 0.1961371 |
| GB50128 | heparanase-like isoform X3 | 11.916400 | -0.1022705 | 0.0322471 | 0.0928074 | 0.2054211 |
| GB42689 | uncharacterized protein LOC100577561 | 11.680716 | -0.4692589 | 0.1292755 | 0.0069494 | 0.2558572 |
| GB46728 | leucine-rich repeat-containing protein 49-like | 11.540281 | -0.0824552 | -0.1745576 | 0.0719110 | 0.0804678 |
| GB55103 | max-like protein X-like | 11.450390 | 0.2397523 | 0.0808834 | -1.3119168 | 0.1406888 |
| GB55817 | uncharacterized protein LOC100577885 | 11.403208 | -0.2780820 | 0.2076758 | -0.0212456 | 0.4120371 |
| GB45828 | uncharacterized J domain-containing protein C4H3.01-like | 11.251023 | -0.3242114 | -0.0523668 | 0.0309067 | 0.1654834 |
| GB45314 | cGMP-dependent 3’,5’-cyclic phosphodiesterase-like isoform 1 | 11.243801 | -0.2591258 | 0.0355666 | -0.0076148 | 0.1053822 |
| GB50168 | uncharacterized protein LOC100576411 | 11.222888 | -0.1188431 | 0.0486892 | 0.3251330 | 0.1152518 |
| 102656107 | immunoglobulin-binding protein 1-like | 11.024003 | -0.0731012 | -0.0155582 | 0.0696120 | 0.2586625 |
| GB55918 | tRNA-splicing endonuclease subunit Sen34-like | 10.938366 | -0.6144494 | 0.1517092 | 0.0806545 | 0.3586086 |
| GB45820 | protoheme IX farnesyltransferase, mitochondrial | 10.935875 | -0.3294710 | -0.0631911 | -0.4850613 | 0.2609807 |
| GB43081 | FAD synthase-like isoform X2 | 10.853091 | -0.2200719 | -0.2625370 | 0.5380586 | 0.3827706 |
| GB40107 | ATPase WRNIP1-like isoform X2 | 10.770622 | -0.0680120 | -0.1284533 | 0.0650551 | 0.2742944 |
| GB55997 | low density lipoprotein receptor adapter protein 1-B-like isoform X1 | 10.749364 | -0.1064426 | -0.1634771 | 0.0377880 | 0.1063477 |
| 102656249 | protein HEXIM1-like | 10.728376 | -0.1017572 | -0.0221028 | 0.0928674 | 0.1261675 |
| GB45215 | peroxisomal N(1)-acetyl-spermine/spermidine oxidase-like | 10.719863 | -0.2253839 | 0.2546455 | 0.5512815 | 0.2428632 |
| GB40874 | GTP-binding protein 1-like isoform X2 | 10.704945 | -0.0723281 | -0.2352593 | 0.0940828 | 0.1700627 |
| GB55259 | caskin-1-like | 10.641186 | -0.2660186 | 0.0175124 | -0.0012038 | 0.1356145 |
| GB47328 | tRNA selenocysteine 1-associated protein 1-like isoform X1 | 10.631021 | 0.1237625 | 0.0612510 | -0.0235768 | 0.1351466 |
| GB41373 | protein germ cell-less isoform X2 | 10.598599 | 0.2034742 | -0.0160647 | 0.1517470 | 0.2301750 |
| 413672 | hexosaminidase D-like isoform 1 | 10.584634 | 0.1916885 | 0.0972348 | 0.0281636 | 0.1812310 |
| GB43421 | sprouty-related, EVH1 domain-containing protein 2 isoform X4 | 10.563881 | -0.1693062 | 0.1874316 | 0.2091097 | 0.1353687 |
| GB55427 | uncharacterized protein LOC100577661 isoform 1 | 10.557185 | -0.2699113 | -0.1622437 | -0.0352591 | 0.3034736 |
| GB49081 | 60 kDa SS-A/Ro ribonucleoprotein-like | 10.551845 | -0.1006043 | -0.0578156 | -0.0594771 | 0.1751285 |
| GB44319 | glycerate kinase-like | 10.551343 | -0.4246135 | -0.1314878 | -1.2225559 | 0.2455242 |
| GB40020 | ras-related protein Rab-39A isoform X1 | 10.494558 | -0.0858439 | -0.0549383 | 0.1668766 | 0.3659913 |
| GB48996 | putative protein arginine N-methyltransferase 10-like isoform X1 | 10.471428 | -0.4255718 | -0.0870829 | 0.1160531 | 0.1658450 |
| GB41067 | dual specificity mitogen-activated protein kinase kinase dSOR1 | 10.427064 | 0.0814680 | 0.2582108 | 0.1385475 | 0.2441206 |
| GB52007 | DCN1-like protein 4-like isoform X1 | 10.391826 | -0.0198666 | 0.0903189 | 0.0216710 | 0.1280059 |
| GB55558 | biogenesis of lysosome-related organelles complex 1 subunit 3-like isoform 1 | 10.325377 | -0.0090096 | -0.0334854 | 0.1398105 | 0.3042514 |
| GB42960 | protein CIP2A-like isoform X1 | 10.182340 | -0.9553921 | 0.0589868 | -0.0068549 | 0.2577863 |
| GB44555 | uncharacterized protein LOC413653 isoformX1 | 10.150862 | -0.0410051 | -0.0667411 | 0.0661551 | 0.1675993 |
| GB41398 | MATH and LRR domain-containing protein PFE0570w-like isoform X1 | 10.107369 | -0.3047154 | -0.1261743 | 0.0283340 | 0.5065219 |
| GB41656 | putative leucine-rich repeat-containing protein DDB_G0290503-like | 10.089747 | -0.7611624 | 0.1408522 | 0.0190563 | 0.2586829 |
| GB51592 | COP9 signalosome complex subunit 2-like | 9.984610 | 0.1667386 | 0.0676922 | -0.0448432 | 0.4956047 |
| GB46813 | unconventional myosin-Ie-like | 9.977965 | -0.0242366 | -1.5771029 | 0.0406426 | 0.0519300 |
| GB54480 | probable RISC-loading complex subunit BRAFLDRAFT_242885 | 9.966272 | -0.6628841 | 0.1009722 | -0.0780035 | 0.1957360 |
| GB51633 | protein HIRA homolog | 9.919911 | 0.3800965 | 0.2041960 | 0.0838411 | 0.1944725 |
| GB54677 | U3 small nucleolar ribonucleoprotein protein IMP3-like | 9.882900 | 0.0178532 | -0.0764870 | 0.0805345 | 0.2872045 |
| GB43483 | golgin-84 | 9.845950 | -0.3196331 | 0.1924280 | 0.1232786 | 0.1407540 |
| GB46387 | zinc finger protein 511-like | 9.697094 | -0.1322206 | -0.1146506 | 0.2697659 | 0.2078972 |
| GB49383 | CUE domain-containing protein 2-like | 9.621938 | 0.4054303 | -0.0980645 | 0.0998646 | 0.3538394 |
| GB55269 | protein PAT1 homolog 1 | 9.615923 | -0.3500669 | 0.0526696 | 0.3860959 | -0.2648133 |
| GB40652 | protein ST7 homolog isoform 1 | 9.587042 | -0.1006532 | 0.2472419 | 0.0872273 | 0.1603035 |
| GB55873 | proteasome inhibitor PI31 subunit-like isoform X1 | 9.569230 | 0.4170462 | -0.0441260 | 0.0671614 | 0.2391310 |
| GB44694 | ras-related protein Rab-43 | 9.546042 | 0.0300555 | 0.0084550 | -0.0173195 | 1.0982301 |
| GB42964 | beta-1,3-glucosyltransferase-like isoform 2 | 9.389693 | 0.1849362 | -0.0012369 | 0.1121248 | 0.2917979 |
| GB43872 | coiled-coil domain-containing protein 50-like | 9.373466 | -0.0204912 | 0.0077092 | 0.1081474 | 0.2352293 |
| GB55872 | DNA repair protein XRCC1-like | 9.360344 | -0.3762419 | -0.0305158 | -0.0193476 | 0.3051661 |
| GB50102 | polyadenylate-binding protein-interacting protein 2 isoform X2 | 9.354473 | 0.2155244 | -0.0221115 | 0.2099256 | 0.3335355 |
| GB53962 | protein 60A | 9.273886 | -0.3128897 | -0.0628346 | 0.0501850 | 0.1949866 |
| GB55722 | G/T mismatch-specific thymine DNA glycosylase-like | 9.253636 | 0.2311669 | 0.2634493 | -0.0252766 | 0.2009277 |
| GB41824 | solute carrier family 25 member 36-A-like isoform X4 | 9.233040 | -0.4829980 | -0.0337348 | 0.0578549 | 0.3871274 |
| GB48579 | zinc finger protein 277-like | 9.136576 | -0.5351122 | 0.2658176 | 0.0521628 | 0.1414883 |
| GB41602 | facilitated trehalose transporter Tret1-like isoform X6 | 9.111198 | 0.1527587 | -0.0235362 | 0.2314605 | 0.0898031 |
| GB46023 | protein PF14_0175-like | 9.106906 | -0.1449085 | 0.0751134 | -0.0575152 | 0.1602656 |
| GB46502 | uncharacterized protein LOC724680 | 9.063368 | 0.2583052 | -0.0240924 | 0.0447010 | -0.4008139 |
| GB49246 | protein tincar isoform X5 | 9.046786 | -0.1626303 | 0.2275856 | 1.4642757 | 0.2101615 |
| GB53932 | protein phosphatase 1L-like isoform X2 | 8.915406 | -0.2084763 | -0.1143118 | 0.8538509 | 0.1344871 |
| GB55535 | E3 ubiquitin-protein ligase Siah1 isoform X1 | 8.906209 | 0.0572880 | -0.1188876 | 0.0221456 | 0.3170472 |
| GB45679 | polycomb protein Asx-like isoform X2 | 8.872479 | -0.4419213 | 0.1477907 | 0.2761911 | 0.5989805 |
| GB55544 | endoplasmic reticulum oxidoreductin-1-like | 8.869211 | 0.3490348 | -0.0378387 | -0.5464275 | 0.1671367 |
| GB49101 | ras-related protein Rab-9A-like isoform X3 | 8.862455 | -0.0206897 | -0.1409925 | 0.2844232 | 0.2345694 |
| GB54838 | post-GPI attachment to proteins factor 2-like isoform X1 | 8.850442 | -0.2082610 | -0.0650075 | 0.1794044 | 0.2041691 |
| GB53648 | tRNA pseudouridine synthase-like 1-like isoform X2 | 8.838015 | -0.1667203 | 0.0819144 | 0.1234332 | 0.1358645 |
| 102654261 | uncharacterized protein C24H6.02c-like | 8.814761 | 0.2002885 | 0.0351338 | 0.0945001 | 0.2413643 |
| GB53315 | uncharacterized protein LOC726215 | 8.770650 | -0.2142003 | -0.1544023 | -0.4663641 | 0.1787594 |
| GB54386 | cyclin-G2 isoform X1 | 8.735376 | -0.1229330 | 0.0194182 | 0.1359141 | 0.1554231 |
| GB49702 | mitoferrin-1 isoformX2 | 8.582951 | -0.4602646 | -0.0066281 | -0.0763098 | 0.2528233 |
| GB40705 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1-like isoform X3 | 8.569997 | -0.2800522 | -0.2407224 | 0.1380000 | 0.2448381 |
| GB43479 | cyclin-C | 8.542879 | 0.4194176 | 0.2036795 | 0.0301399 | 0.3590183 |
| GB49429 | calcium channel flower-like isoform X1 | 8.540568 | 0.5949925 | 0.0622683 | 0.1972320 | 0.2291893 |
| GB48358 | chromosome transmission fidelity protein 8 homolog | 8.532017 | 0.1553744 | 0.2128898 | 0.0886741 | 0.4512296 |
| GB53702 | uncharacterized protein C45G9.7-like | 8.529210 | -0.8796477 | 0.0639904 | -0.0229375 | 0.1756738 |
| GB44365 | germ cell-expressed bHLH-PAS-like protein, transcript variant X3 | 8.523907 | -0.3566238 | -0.1493375 | 0.0838529 | 0.1391885 |
| GB55564 | transcriptional regulator ATRX homolog isoform X2 | 8.513158 | 0.1657669 | 0.0757655 | 0.0806177 | 0.2229550 |
| GB47249 | E3 ubiquitin-protein ligase Smurf1 isoform X2 | 8.446508 | 0.3279116 | 0.0698081 | 0.2139638 | 0.2055806 |
| GB54842 | arginine–tRNA ligase, cytoplasmic | 8.442742 | -0.1656897 | 0.0703201 | -0.0895508 | 0.1622339 |
| GB51498 | myeloid differentiation primary response protein MyD88-A isoform X1 | 8.411756 | -0.6845852 | -0.0370699 | -0.0431235 | 0.1189769 |
| GB46148 | guanine nucleotide-binding protein subunit alpha homolog | 8.391180 | -0.5698858 | 0.1889987 | -0.0114847 | 0.2591016 |
| GB50834 | peroxisomal membrane protein 11B-like isoform 2 | 8.369996 | -0.3418630 | -0.0181533 | 0.0621884 | 0.2242913 |
| GB43388 | transcription initiation factor IIA subunit 2 | 8.153357 | 0.3747687 | -0.0179152 | 0.0932493 | 0.2690722 |
| GB44404 | retinol dehydrogenase 13-like | 8.146156 | 0.4662618 | 0.0892959 | -1.5786809 | 0.2339449 |
| GB45142 | RB1-inducible coiled-coil protein 1 isoform X1 | 8.108184 | -1.0065398 | -0.0271733 | 0.1124220 | 0.2110064 |
| GB46745 | general transcription factor 3C polypeptide 3-like | 8.092430 | 0.2195164 | 0.0938966 | 1.0632004 | 0.1579925 |
| GB50255 | RWD domain-containing protein 1-like isoform X1 | 8.092190 | 0.1247443 | -0.0452487 | 0.5446836 | 0.3149420 |
| GB48642 | uncharacterized protein LOC100577967 | 8.013801 | -0.6570737 | -0.3268260 | 0.0729872 | 0.2644375 |
| GB51251 | ADP-ribosylation factor-like protein 8B-A-like isoform X1 | 8.011143 | 0.4567300 | 0.0733491 | 0.1276877 | 0.2459041 |
| 102653960 | magnesium-dependent phosphatase 1-like | 7.934631 | -0.2650674 | -0.1497835 | -0.0683272 | 0.2441625 |
| GB44697 | probable serine/threonine-protein kinase DDB_G0283337-like isoform X2 | 7.878991 | -0.4302061 | 0.4202618 | 0.1557601 | 0.1325223 |
| GB55038 | protein UXT homolog | 7.873234 | 0.3031172 | -0.0474639 | 0.0250510 | 0.2643234 |
| GB54108 | dual specificity protein phosphatase 3-like isoform X2 | 7.871551 | 0.0337993 | -0.0415448 | 0.0811359 | 0.1768748 |
| GB42078 | uncharacterized protein LOC725568 | 7.845221 | 0.3982550 | 0.0297880 | 0.2463155 | 0.3734292 |
| GB48617 | uncharacterized protein C15orf41 homolog isoform X1 | 7.838868 | 0.1648228 | 0.1994837 | 0.0422280 | 0.2866344 |
| 100577724 | ELMO domain-containing protein 2-like isoform X2 | 7.791314 | -0.1282071 | 0.0771536 | 0.1802733 | 0.1810349 |
| GB55523 | LOW QUALITY PROTEIN: leucine-rich repeats and immunoglobulin-like domains 3 | 7.789942 | 0.2428644 | 0.0893082 | 0.1680035 | 0.1281048 |
| GB56016 | 39S ribosomal protein L30, mitochondrial | 7.754830 | 0.0139581 | -0.0721010 | 0.1219559 | 0.1989394 |
| GB55191 | uncharacterized protein LOC100576289 | 7.708494 | -1.0449993 | 0.2183642 | 0.2056562 | 0.4601299 |
| 102656444 | protein PFC0760c-like isoform X1 | 7.681926 | -1.3055158 | 0.0315297 | 0.1198871 | 0.1882784 |
| GB46146 | sodium/potassium-transporting ATPase subunit beta-2 | 7.674605 | 0.5096384 | 0.0596106 | 0.2390689 | 0.3285983 |
| GB45376 | putative peptidyl-prolyl cis-trans isomerase dodo | 7.612748 | 0.3745301 | 0.0839339 | 0.1322601 | 0.2814442 |
| GB55587 | OTU domain-containing protein 7B-like isoform X2 | 7.608980 | 0.0299225 | -0.0545040 | 0.1380641 | 0.3242408 |
| GB48809 | proline-, glutamic acid- and leucine-rich protein 1-like | 7.579596 | 0.0611634 | 0.0002520 | -0.8671498 | 0.1492251 |
| GB41208 | cell division control protein 45 homolog isoform X2 | 7.518906 | 0.0962735 | 0.0836906 | 0.0447179 | 0.2958022 |
| GB49178 | OTU domain-containing protein 6B-like | 7.516930 | -0.0953920 | 0.0173889 | 0.0382644 | 1.5062191 |
| GB50984 | sorting nexin-17 isoform X2 | 7.515664 | -0.4549651 | -0.2277571 | 0.0963375 | 0.0739402 |
| 102656287 | transmembrane protein 216-like | 7.509296 | 0.1906785 | 0.0399544 | 0.0716641 | -0.0022660 |
| GB41970 | ras-like protein 2-like isoform X1 | 7.433796 | -0.0351236 | -0.1258498 | 0.0709487 | 0.2934914 |
| GB51623 | adenylate kinase isoenzyme 6 isoform X2 | 7.399046 | 0.0468456 | 0.0217309 | 0.1080657 | 0.0880704 |
| GB52210 | DDB1- and CUL4-associated factor 12-like isoform X3 | 7.313239 | -0.1762286 | -0.0753000 | 0.2127309 | 0.1706834 |
| 102655090 | uncharacterized protein LOC102655090 | 7.265020 | -0.6513193 | 0.2082596 | 0.0683780 | -0.0674814 |
| GB49727 | prostaglandin E2 receptor EP4 subtype-like isoform X3 | 7.215886 | -0.4599105 | -0.1748025 | -0.3188992 | 0.4054800 |
| GB54267 | serendipity locus protein H-1-like | 6.903789 | 0.3323666 | -0.0811215 | 0.1738090 | 0.3864286 |
| GB47475 | protein lethal(2)essential for life-like isoform 1 | 6.295748 | 0.2576099 | 0.1582286 | 0.5862057 | 0.3050397 |
| GB50748 | PAX3- and PAX7-binding protein 1-like | 5.296029 | -0.1356224 | 0.0316976 | 0.0392064 | -0.2173359 |
| GB49188 | intraflagellar transport protein 80 homolog isoform X3 | 5.170123 | -0.1611611 | -0.5360297 | 0.9444551 | 0.2601014 |
| GB40147 | fez family zinc finger protein 1-like | 2.799368 | -0.3006173 | 0.3022265 | 0.6688060 | 0.3775222 |
Module 7
Table S32: List of all the genes in Module 7, ranked by their within-module connectivity, k. The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
gene_list <- module_gene_list(7)
saveRDS(gene_list, file = "supplement/tab_S32.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB43212 | ubiquitin-protein ligase E3C-like isoform X2 | 19.056059 | 0.2526930 | -0.0948782 | 0.0449179 | -0.0295972 |
| GB43857 | importin-13 isoform X1 | 18.128184 | 0.1389376 | -0.0024907 | 0.0379327 | -0.0828945 |
| GB53778 | huntingtin-like | 18.037608 | 0.0668536 | -0.0653910 | 0.4847436 | 0.0275824 |
| GB47281 | kinesin heavy chain isoform 1 | 17.614981 | 0.0268912 | -0.0719550 | -0.5362209 | -0.2925697 |
| GB40720 | CCR4-NOT transcription complex subunit 3-like isoform X1 | 15.793193 | 0.1047626 | 0.1316095 | 0.0452534 | 0.3251685 |
| GB41872 | ATP-binding cassette sub-family F member 3-like isoform X2 | 15.353997 | 0.1512023 | 0.2038624 | -0.7581738 | -0.0150930 |
| GB49120 | raf homolog serine/threonine-protein kinase phl | 14.819233 | -0.1251284 | -0.0020563 | 0.1969402 | -0.0230406 |
| GB41128 | ubiquitin thioesterase OTU1-like | 14.323821 | 0.1977835 | 0.0352611 | -0.0854440 | -0.1282436 |
| GB50061 | uncharacterized protein LOC410606 | 14.205891 | -0.0230116 | -0.0157831 | 0.3028269 | 0.0531771 |
| GB44884 | splicing factor 3B subunit 3 isoform 1 | 14.125689 | 0.3007990 | 0.0352040 | 0.0678768 | -0.0398547 |
| GB53220 | ubiquitin carboxyl-terminal hydrolase 3-like isoform X1 | 14.004349 | 0.1073774 | 0.0353565 | 0.0968747 | -0.0846290 |
| GB55770 | TBC1 domain family member 9 | 13.727593 | 0.0028834 | 0.0039453 | 0.1252382 | 0.1095110 |
| GB46321 | neurofibromin isoform X1 | 13.682566 | 0.2333111 | -0.1681329 | 1.2220114 | -0.0811956 |
| GB54557 | arginine/serine-rich protein PNISR-like isoform X2 | 13.592683 | 0.0855084 | 0.1182126 | 0.1386842 | 0.0896055 |
| GB42838 | RNA-binding protein 39-like isoform X5 | 13.592240 | -0.0858264 | -0.0235484 | 0.1438511 | 0.0867029 |
| GB43304 | cadherin-87A-like isoform X1 | 13.522761 | 0.0414844 | 0.0561924 | 0.6879037 | 0.0415333 |
| GB42436 | protein arginine N-methyltransferase 7-like isoform X2 | 13.502118 | 0.0458574 | -0.1217573 | 0.0691762 | 0.0793897 |
| GB50214 | uncharacterized protein LOC409502 | 13.342240 | 0.1933261 | 0.0493912 | 0.3439567 | -0.0386244 |
| GB49152 | TBC1 domain family member 24-like isoformX1 | 13.204631 | -0.1030115 | 0.0041823 | 0.0527562 | 0.0346674 |
| GB43236 | probable tRNA (uracil-O(2)-)-methyltransferase-like | 13.092529 | -0.0304216 | 0.3187739 | 0.0580359 | -0.0120557 |
| GB53708 | serine/threonine-protein kinase unc-51 | 13.071398 | 0.2529429 | 0.0097991 | -0.2992022 | 0.0472491 |
| GB50725 | vang-like protein 2-like isoform X2 | 13.068606 | 0.1737603 | 0.0114350 | 0.1756634 | 0.0752588 |
| GB54230 | uncharacterized protein LOC551498 | 13.029864 | 0.0892138 | 0.0792454 | 0.8939247 | 0.1081059 |
| GB44899 | trithorax group protein osa isoform X6 | 12.694162 | -0.2152737 | -0.0501742 | 0.1631650 | -0.1390978 |
| GB44490 | hornerin | 12.686557 | 0.0804526 | -0.0218843 | 0.1554077 | -0.0109212 |
| GB47322 | glycine-rich cell wall structural protein 1.8-like isoform X10 | 12.632665 | 0.1452844 | 0.1620032 | 0.4793091 | -0.0500198 |
| GB43467 | rho guanine nucleotide exchange factor 28-like isoform X8 | 12.585898 | 0.0956703 | -0.0501709 | 0.0950157 | -0.0466181 |
| GB54590 | polyadenylate-binding protein 1-like isoform X2 | 12.503830 | 0.4827100 | 0.1573794 | 0.1675415 | -0.1885244 |
| GB45972 | neural-cadherin isoform X2 | 12.370171 | -0.1470460 | 0.0024069 | 1.3812275 | -0.0183059 |
| 100576876 | intracellular protein transport protein USO1-like isoform X1 | 12.313808 | -0.0128192 | -0.0632411 | 0.1493875 | -0.0754738 |
| GB44422 | uncharacterized protein LOC412543 isoform X3 | 12.033349 | 0.6015819 | -0.0529977 | -0.0100398 | 0.0537208 |
| GB40676 | phosphatidylinositol 4-kinase beta-like | 11.710006 | 0.1395238 | 0.0100279 | 0.1130146 | -0.0094421 |
| GB54731 | plasmolipin-like isoform 1 | 11.623575 | 0.4047489 | 0.0057446 | 0.0648378 | -0.0065488 |
| GB44761 | Rab GTPase activating protein 10 isoform X3 | 11.325687 | 0.0665230 | 0.0331507 | 0.1325785 | -0.0485636 |
| GB50373 | endophilin-A isoform X1 | 11.222091 | 0.1707438 | 0.1446588 | 0.1530334 | -0.0721249 |
| 102655673 | mitochondrial import inner membrane translocase subunit Tim16-like | 11.154858 | -0.0848364 | 0.0642094 | 0.2513499 | 0.0855126 |
| GB49375 | homeobox protein PKNOX2-like isoform X1 | 10.805109 | -0.1368990 | 0.0571144 | 0.1485631 | 0.0049030 |
| GB41423 | dosage compensation regulator isoform X3 | 10.678731 | 0.0680768 | 0.0153003 | 0.1111923 | -0.0725054 |
| GB44419 | protein peanut isoform X3 | 10.578227 | 0.2559230 | 0.1256845 | 0.1371956 | -0.2211204 |
| GB43899 | E3 ubiquitin-protein ligase MIB2-like isoform X2 | 10.573502 | 0.0424029 | 0.1632035 | 0.0911580 | 0.0159632 |
| GB44259 | aryl hydrocarbon receptor nuclear translocator homolog isoform X3 | 10.557091 | 0.1265462 | 0.1097368 | 0.2360432 | 0.0212316 |
| GB52106 | tolkin isoform X1 | 10.503840 | 0.4308379 | -0.2027848 | 0.1068198 | -0.1203723 |
| GB48337 | protein crooked neck | 10.416603 | 0.0203727 | 0.1215738 | 0.0384466 | 0.0818576 |
| GB49960 | uncharacterized protein LOC100379261 | 10.136789 | 0.1783426 | -0.1747512 | 0.0957692 | -0.0287478 |
| GB55574 | probable phospholipid-transporting ATPase VD-like, transcript variant X3 | 10.031602 | -0.1535478 | 0.0833423 | 0.1986956 | 0.0493337 |
| GB44679 | metastasis-associated protein MTA3 isoform X2 | 9.907001 | 0.0122040 | 0.1672956 | 0.4215435 | -0.0436046 |
| GB51337 | dedicator of cytokinesis protein 3-like isoform X2 | 9.890689 | -0.0351151 | -0.0855787 | 0.2481693 | -0.4953984 |
| GB55162 | uncharacterized protein LOC551144 isoform X1 | 9.833976 | 0.1580812 | 0.0152496 | 0.1873342 | 0.0159339 |
| GB53787 | lysine-specific demethylase 3B-like isoform X4 | 9.830941 | -0.2273710 | 0.0249965 | -0.1374384 | -0.4127317 |
| GB52510 | putative uncharacterized protein DDB_G0277255 isoform X2 | 9.723553 | 0.0343818 | 0.0289312 | -0.0529365 | -0.1318499 |
| GB45414 | nuclear hormone receptor FTZ-F1 beta isoform X1 | 9.714424 | -0.0053048 | -0.1307936 | 0.2488088 | 0.0365510 |
| GB50344 | beta-1,4-mannosyltransferase egh | 9.702963 | 0.2559145 | -0.1136370 | 0.2247666 | 0.0998973 |
| GB52075 | RNA-binding protein 45-like isoform X1 | 9.609042 | 0.1871956 | 0.1231640 | 0.5239350 | -0.0950067 |
| GB48932 | phosphoinositide 3-kinase regulatory subunit 4 isoform X2 | 9.557524 | 0.0028169 | -0.0453824 | 0.0006408 | -0.0518159 |
| GB51219 | eye-specific diacylglycerol kinase isoform X2 | 9.487613 | -0.1059247 | 0.3389632 | 0.6697580 | -0.0121393 |
| 409956 | uncharacterized protein LOC409956 isoform X2 | 9.483005 | 0.0560777 | -0.0878618 | 0.1549100 | -0.3010497 |
| GB49505 | rho-related BTB domain-containing protein 1 isoform X3 | 9.476015 | 0.0573503 | -0.1398313 | 0.1267769 | 0.0135867 |
| GB47816 | splicing factor 1-like isoform X2 | 9.466436 | 0.2446155 | 0.0839599 | 0.5098015 | 0.0679864 |
| GB51841 | multiple inositol polyphosphate phosphatase 1-like | 9.400309 | 0.0637724 | 0.0183885 | 0.2287712 | -0.0324923 |
| GB54551 | slit homolog 3 protein-like isoform X2 | 9.332628 | 0.3282476 | 0.0479686 | 0.3510590 | 0.0653779 |
| GB55323 | phosphatidylinositol 4-phosphate 5-kinase type-1 gamma isoform X8 | 9.311576 | 0.2320622 | -0.0964140 | 0.2496250 | -0.0139458 |
| GB44412 | protein turtle homolog A-like | 9.141434 | 0.3208528 | 0.1792722 | 0.2424071 | -0.1619378 |
| GB49921 | integrin alpha-PS2 isoform X1 | 9.125215 | 0.0711565 | -0.1290202 | 0.1068253 | 0.0413718 |
| 726252 | titin-like isoform X2 | 9.120307 | 0.1557374 | -0.0685572 | 0.1987358 | 0.0065025 |
| GB46271 | protein BCL9 homolog isoform X1 | 9.051295 | -0.1609479 | 0.0074360 | -1.6494149 | -0.0518614 |
| GB45593 | zinc finger protein Helios-like | 9.040495 | 0.0951445 | -0.0285509 | 0.1549917 | -0.0961719 |
| GB47028 | enoyl-CoA hydratase domain-containing protein 3, mitochondrial-like | 9.008486 | 0.5278010 | -0.0631981 | 0.1353259 | -0.0192182 |
| GB44534 | spastin isoform X1 | 9.005178 | 0.4578382 | -0.0429820 | 0.0587009 | -0.0112236 |
| GB44779 | tyrosine-protein phosphatase non-receptor type 4 isoform X2 | 8.952552 | 0.0141231 | 0.1514778 | 0.0333931 | 0.0462535 |
| GB51209 | dentin sialophosphoprotein-like isoform X4 | 8.940775 | -0.4228685 | -0.0171232 | 0.1264229 | -0.1038103 |
| 102654691 | protein translation factor SUI1 homolog | 8.922963 | 0.4450016 | -0.0120966 | 0.6470182 | 0.0314599 |
| 102655836 | tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6-like | 8.899108 | 0.1909867 | -0.0206296 | 0.0340970 | -0.1561803 |
| GB52185 | uncharacterized protein LOC100578041 isoformX1 | 8.889372 | 0.2006631 | -0.1834607 | 0.2037535 | 0.0035903 |
| GB41225 | aquaporin AQPAn.G-like isoform X2 | 8.854883 | 0.1540005 | -0.3514298 | 0.1776022 | 0.4935015 |
| GB52779 | probable cation-transporting ATPase 13A3-like isoform X3 | 8.695500 | -0.2401490 | 0.0967486 | 0.0578498 | -0.1343988 |
| GB45955 | beta-galactosidase-like isoform X2 | 8.668723 | 0.5822940 | 0.0651901 | 0.2149433 | -0.1643084 |
| GB53592 | SNF-related serine/threonine-protein kinase-like | 8.654279 | 0.2903133 | 0.1083014 | 0.2636373 | 0.5191470 |
| GB52157 | GRAM domain-containing protein 3-like isoform X2 | 8.647084 | 0.2922811 | -0.1182924 | -0.8010004 | 0.0652401 |
| GB47669 | putative uncharacterized protein DDB_G0271606-like | 8.568163 | 0.2204971 | -0.1173649 | -0.1299728 | 0.0992965 |
| GB40417 | transmembrane protein 98 | 8.560464 | 0.4995915 | 0.0023776 | 0.1333669 | -1.4896202 |
| GB49911 | insulin receptor substrate 1 isoform X3 | 8.484400 | 0.0986907 | -0.1734225 | 0.3751177 | -0.0295494 |
| GB47918 | netrin receptor UNC5C isoform X8 | 8.461620 | 0.2401296 | 0.0931734 | 0.1131221 | -0.1243948 |
| GB53437 | F-box only protein 32-like isoform X2 | 8.350333 | 0.1910897 | -0.1648703 | 0.2951614 | 0.0542353 |
| GB46918 | monocarboxylate transporter 13-like, transcript variant X2 | 8.342444 | 0.4279166 | 0.1059473 | 0.2234353 | 0.0141015 |
| GB42976 | teneurin-a-like isoform X7 | 8.326042 | 0.3393374 | -0.0503122 | 0.9972151 | 0.3059716 |
| 409007 | heterogeneous nuclear ribonucleoprotein K isoform X8 | 8.271593 | -0.0604939 | -0.0706296 | 0.4540430 | -0.3359263 |
| GB50892 | uncharacterized protein LOC100577980 isoform X3 | 8.195928 | 0.1032354 | -0.3067119 | -0.1367296 | -0.1184458 |
| GB42979 | ankyrin repeat and BTB/POZ domain-containing protein BTBD11-like isoform X3 | 8.191034 | 0.3054026 | -0.1904184 | 0.4129782 | 0.0100772 |
| GB54219 | BAG domain-containing protein Samui-like isoform X3 | 8.182365 | -0.1837839 | 0.4111713 | 0.2818847 | -0.1482710 |
| GB51674 | 26S proteasome non-ATPase regulatory subunit 10-like | 8.110245 | 0.2303124 | 0.0276688 | 0.0103197 | -0.0349071 |
| GB54395 | uncharacterized protein LOC413385 | 8.101636 | 0.0895723 | 0.0708472 | 0.0168679 | -0.1011838 |
| GB44968 | metabotropic glutamate receptor 1 | 8.095985 | -0.2838043 | 0.1511773 | 0.2142725 | -0.0724188 |
| GB51744 | uncharacterized protein LOC724439 | 8.091537 | 0.8112050 | -0.2579550 | 0.2886800 | 0.0712415 |
| 724450 | uncharacterized protein LOC724450 isoform X3 | 7.965021 | -0.0063056 | -0.0381084 | 0.2221333 | -0.2398677 |
| GB40162 | chondroitin sulfate synthase 1-like isoform X2 | 7.952768 | 0.1937082 | 0.2118247 | 0.2261672 | -0.3507220 |
| GB40907 | putative ferric-chelate reductase 1 homolog isoform X3 | 7.934787 | 0.1220467 | 0.0794329 | 0.0786812 | -0.2194966 |
| 726866 | uncharacterized protein LOC726866 | 7.922809 | 0.2773045 | 0.0704480 | 0.2425601 | -0.0854166 |
| GB42196 | thrombospondin type-1 domain-containing protein 4-like isoform X3 | 7.898138 | -0.1542267 | -0.0954962 | 0.0678213 | -0.0191430 |
| GB43567 | MAM and LDL-receptor class A domain-containing protein C10orf112-like | 7.893733 | 0.4417807 | -0.0191397 | 0.2231429 | -0.0939693 |
| 413366 | homeobox protein SIX2-like isoform X4 | 7.889655 | -0.8301995 | -0.1007038 | 0.2913582 | -0.1269705 |
| GB42035 | myosin-I heavy chain isoform X2 | 7.882072 | -0.0014280 | 0.3338154 | 0.4125285 | 0.1308000 |
| GB47029 | uncharacterized protein LOC724558 | 7.859954 | 0.6393499 | -0.0504088 | -0.1530074 | 0.0202752 |
| GB42654 | leucine carboxyl methyltransferase 1-like | 7.828984 | 0.2608335 | 0.0339394 | 0.1603670 | -0.2844500 |
| GB52636 | cell growth regulator with RING finger domain protein 1-like isoform X2 | 7.761509 | 0.2801670 | -0.0432581 | 0.1460809 | -0.2455172 |
| 725183 | probable palmitoyltransferase ZDHHC24-like isoform X1 | 7.760804 | 0.0182415 | -0.2681787 | 0.2905207 | -0.0387823 |
| GB43882 | alpha-mannosidase 2 isoform X3 | 7.672613 | 0.4326024 | -0.3898568 | 0.2543905 | -0.0826037 |
| GB51385 | nephrin-like isoform 1 | 7.638084 | -0.3216389 | 0.2435750 | 0.1870275 | 0.0383706 |
| GB52082 | LOW QUALITY PROTEIN: sn1-specific diacylglycerol lipase alpha | 7.609801 | 0.0989794 | 0.0438915 | 0.2350143 | 0.0361958 |
| GB52986 | mitochondrial import inner membrane translocase subunit Tim23-like isoform X1 | 7.587191 | 0.2452917 | 0.0085709 | 0.1445813 | -0.0062553 |
| 102653601 | zinc finger protein 729-like | 7.550321 | 0.0535043 | 0.0075084 | 0.2508535 | -0.3351553 |
| GB46371 | tyrosine-protein kinase Src64B-like isoform X3 | 7.532158 | -0.0663805 | 0.1291985 | 0.2322886 | 0.2277438 |
| GB54032 | methyltransferase-like protein 9-like isoform X2 | 7.483067 | 0.3721757 | 0.1988250 | 0.0772659 | -0.1572309 |
| GB40928 | tripartite motif-containing protein 2-like isoform X1 | 7.480566 | -0.1898551 | -0.0932847 | 0.1595720 | 0.0919059 |
| 102656070 | uncharacterized protein LOC102656070 | 7.385732 | 0.0774192 | -0.3793332 | 0.4132221 | -0.1639907 |
| GB44315 | monocarboxylate transporter 9-like isoform X1 | 7.364144 | 0.4478351 | 0.2227958 | 0.1933958 | 0.1172661 |
| GB42377 | protein giant-lens-like | 7.279187 | 0.0687812 | -0.0370822 | 0.0926618 | -0.0032676 |
| GB44984 | U5 small nuclear ribonucleoprotein 40 kDa protein-like isoform X1 | 7.152506 | 0.5036272 | 0.0632588 | 0.1517729 | -0.0270656 |
| GB42487 | calpain-C isoform X2 | 7.061177 | 0.2641116 | 0.2357378 | 0.1865473 | -0.0532727 |
| GB44041 | dachshund homolog 2-like isoform X3 | 6.949928 | 0.2449004 | -0.0547904 | 0.1286984 | -0.0985249 |
| GB44060 | centrosomal protein of 104 kDa-like isoform X2 | 6.787552 | 0.9893042 | -0.1005035 | 0.2224712 | 0.0542083 |
| GB46749 | endochitinase-like isoform X1 | 6.698633 | 0.6803442 | 0.1809386 | 0.1076001 | -0.0166939 |
| GB42326 | glutamyl aminopeptidase-like isoform X3 | 6.665332 | -0.1918426 | -0.2267862 | -0.1909795 | -0.0729451 |
| GB40531 | uncharacterized protein LOC100578051 isoform X3 | 6.581381 | -0.1778554 | -0.0961652 | 0.2405992 | -0.3229043 |
| 102655815 | suppressor protein SRP40-like | 6.410201 | 0.8524923 | 1.3833606 | 0.8196561 | -0.6248925 |
| GB53146 | uncharacterized protein LOC412149 isoform X1 | 6.401826 | -0.0825530 | 0.1045735 | -0.1856001 | 0.0619452 |
| GB46073 | uncharacterized protein LOC551865 isoformX2 | 6.287155 | -0.0172652 | 0.2258487 | -0.0096061 | 0.0229668 |
| GB43015 | myocardin-related transcription factor A-like isoform X6 | 6.152442 | -0.0367942 | 0.0143781 | 0.2631757 | 0.0627405 |
| 102654007 | uncharacterized protein LOC102654007 | 5.894278 | 0.2417289 | -0.1293139 | 0.2077469 | -0.0148285 |
| 102656939 | histone-lysine N-methyltransferase SETMAR-like | 5.865772 | 0.1497729 | 0.2369577 | 1.0620095 | 0.6002907 |
| GB41647 | transcription factor Sox-7-like isoform X2 | 5.795569 | 0.0244209 | -0.2801421 | 1.4666012 | -0.3879830 |
| GB49810 | RIB43A-like with coiled-coils protein 1-like | 5.283242 | -0.4749690 | -0.1025304 | -0.1589753 | -0.2393070 |
| GB44616 | EF-hand domain-containing family member C2-like isoform X1 | 5.222967 | 0.2363270 | 0.0454848 | 0.0700841 | -0.2935181 |
| GB50734 | uncharacterized protein LOC725625 | 4.955965 | 0.5723249 | -0.2713737 | 0.3683741 | -0.0261181 |
| GB53798 | esterase E4-like | 4.763677 | 0.9132871 | -1.1903127 | 0.2569342 | -0.0940903 |
| GB45427 | Krueppel homologous protein 1 | 4.573111 | -0.0174895 | 0.0395830 | 0.4532421 | -0.2188543 |
| GB42800 | protein takeout-like isoform X1 | 4.225151 | 0.7882925 | 0.1989128 | 0.0197007 | -0.1246820 |
| GB46290 | acetyl-coenzyme A synthetase-like | 4.153359 | 0.1907309 | -0.1625415 | 0.4246826 | -0.2051017 |
| GB44976 | ataxin-2 homolog isoform X3 | 4.149661 | 0.0469414 | -0.0680298 | 0.2666148 | -0.1835732 |
| GB40495 | zinc finger protein 43-like isoform X1 | 3.868333 | 0.5265820 | -0.3355168 | 0.6345599 | -0.0172255 |
| GB40554 | G1/S-specific cyclin-D2 | 3.865592 | 0.1362646 | 0.0943557 | 0.1991426 | -0.3526045 |
| GB45382 | intraflagellar transport protein 74 homolog | 3.715030 | 0.0360060 | -0.1247186 | 0.2448515 | -0.0526646 |
| GB43604 | uncharacterized protein LOC725033 isoform X4 | 3.676268 | 0.3453212 | -0.0200476 | 0.4718334 | 0.1075488 |
| GB51657 | uncharacterized protein LOC100578157 | 3.637955 | 0.1322720 | -0.5335299 | 0.7834386 | -0.3924988 |
| GB41844 | ATP-binding cassette sub-family G member 5-like | 3.372551 | 0.7657014 | 0.1292601 | 0.8080343 | -0.1644371 |
| GB51376 | serotonin receptor | 3.059578 | 1.6897679 | 0.4837407 | 0.1477586 | 0.6097080 |
| GB17921 | dopamine receptor 2 | 2.761384 | 0.2831209 | -0.2845336 | -0.4048745 | -0.0826887 |
| GB45956 | putative succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial-like | 2.708942 | -0.2031321 | -0.2797345 | 0.5862043 | 0.0897216 |
| 102655054 | protein tyrosine phosphatase domain-containing protein 1-like isoform X2 | 2.503884 | 0.8503692 | 0.3750205 | 0.6810604 | 0.1281549 |
| GB50186 | uncharacterized protein LOC100577394 isoform X3 | 2.177106 | 0.0310930 | 0.2985490 | -0.7082269 | 0.2091186 |
| GB46310 | cuticular protein 17 precursor | 1.920129 | 0.2959369 | 0.4322316 | 0.3721362 | -0.1581453 |
Module 8
Table S33: List of all the genes in Module 8, ranked by their within-module connectivity, k. The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
gene_list <- module_gene_list(8)
saveRDS(gene_list, file = "supplement/tab_S33.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB41139 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | 4.4012470 | -0.1191108 | -0.1058419 | 0.0287706 | 0.1583752 |
| GB54940 | growth hormone-inducible transmembrane protein-like isoform X5 | 4.3387588 | 0.0164674 | 0.0684403 | 0.0735684 | 0.0525995 |
| GB46440 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial-like | 4.1000841 | -0.1432112 | -0.0892090 | 0.1102687 | 0.1659014 |
| GB54961 | uncharacterized protein LOC725712 isoform X3 | 3.9219655 | -0.1044514 | -0.0869262 | 0.1242401 | 0.1347024 |
| GB54596 | cytochrome b-c1 complex subunit 7-like | 3.9217965 | -0.1692614 | -0.3258486 | -0.0085974 | 0.1781197 |
| GB49313 | voltage-dependent anion-selective channel | 3.7559077 | 0.1206381 | -0.0351029 | 0.0647365 | 0.1324141 |
| GB55708 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial-like | 3.7328153 | -0.0799560 | -0.1651344 | 0.0902151 | 0.1590548 |
| GB42422 | ADP/ATP translocase | 3.5940991 | -0.0367367 | -0.2829944 | 0.0541088 | 0.1539769 |
| GB46369 | cytochrome c1, heme protein, mitochondrial isoform X3 | 3.5878684 | -0.0542768 | -0.0884751 | 0.3460644 | 0.1706220 |
| GB42929 | cytochrome c oxidase subunit 4 isoform 1, mitochondrial isoform X1 | 3.5862599 | 0.0085438 | -0.2688416 | 0.1296427 | 0.1435245 |
| GB41741 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mitochondrial-like isoform X2 | 3.5735884 | -0.0847110 | -0.1714775 | 0.1331272 | 0.1832255 |
| GB43629 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-like | 3.5194076 | 0.0958536 | -0.1643438 | 0.1581970 | 0.2637005 |
| GB41028 | ATP synthase subunit alpha, mitochondrial isoform 1 | 3.5072605 | -0.0553949 | -0.0931082 | 0.2360865 | 0.3746952 |
| GB52753 | succinate dehydrogenase cytochrome b560 subunit, mitochondrial | 3.4853499 | 0.0810905 | -0.1775820 | 0.2742764 | 0.2041456 |
| GB53749 | cytochrome c oxidase subunit 6A1, mitochondrial | 3.4696997 | 0.1283259 | -0.2946381 | 0.1176364 | 0.1801702 |
| GB48784 | cytochrome c | 3.4587225 | -0.0042056 | -0.3979038 | 0.2745145 | 0.7787113 |
| GB46882 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | 3.4285793 | 0.0165555 | -0.0229026 | 0.1400815 | 0.5987601 |
| GB55643 | ATP synthase subunit O, mitochondrial | 3.3805937 | -0.1221369 | -0.3889205 | 0.1422731 | 0.1747252 |
| GB47679 | putative ATP synthase subunit f, mitochondrial-like | 3.3712761 | 0.0832491 | -0.1522548 | 0.1173262 | 0.1055939 |
| GB52736 | ATP synthase subunit beta, mitochondrial isoform X1 | 3.3560844 | -0.0294307 | -0.0033689 | 0.2970809 | -0.0057924 |
| GB47500 | mitochondrial-processing peptidase subunit beta-like | 3.3452725 | -0.1881842 | -0.0597179 | 0.0811601 | 0.2026737 |
| GB51086 | ATP synthase subunit delta, mitochondrial isoform 3 | 3.3406699 | -0.0674897 | -0.0753343 | 0.1463538 | 0.2702057 |
| GB41143 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | 3.2972912 | -0.3013049 | -0.1902631 | 0.1367286 | 0.1111649 |
| GB47886 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7-like | 3.2817339 | 0.1186647 | -0.1424645 | 0.2245911 | 0.1801420 |
| GB51192 | cytochrome b-c1 complex subunit Rieske, mitochondrial | 3.2074650 | 0.0374775 | -0.1043468 | 0.1835974 | 0.6273204 |
| GB54687 | phosphate carrier protein, mitochondrial-like isoform 1 | 3.1946397 | 0.0437169 | -0.2881002 | 0.0773879 | 0.1222814 |
| GB49306 | ATP synthase subunit gamma, mitochondrial isoform X1 | 3.1353414 | -0.2494563 | -0.1097028 | -0.1395306 | -0.1783887 |
| GB43704 | adenylosuccinate synthetase-like | 3.0832059 | 0.0505546 | -0.0945907 | 0.1562562 | 0.1608233 |
| GB45153 | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial isoform X2 | 3.0486283 | -0.0544827 | -0.0287016 | 0.3253613 | 0.1114881 |
| GB45099 | superoxide dismutase 2, mitochondrial | 3.0363301 | -0.1393383 | -0.2225523 | 0.0754449 | 0.1540678 |
| GB44608 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-like isoform 1 | 2.7112823 | 0.1746420 | 0.0616242 | 0.1065548 | 0.1389951 |
| GB50918 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | 2.6924420 | -0.2445495 | -0.1469059 | 0.0607883 | 0.0368726 |
| GB50554 | negative elongation factor A-like | 2.5858228 | 0.2354063 | 0.0531283 | 0.0691260 | 0.1634791 |
| GB42871 | putative ATP-dependent Clp protease proteolytic subunit, mitochondrial-like isoform X4 | 2.2253988 | -0.0140272 | -0.0483964 | 0.0730049 | 0.1478795 |
| GB43248 | alpha glucosidase 2 precursor | 2.1469572 | 0.1316442 | 0.1323163 | -0.2584368 | -0.1531523 |
| GB50268 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial isoform 2 | 2.0872991 | -0.1972878 | -0.0736731 | 0.1255290 | 0.2146555 |
| GB45731 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | 2.0770835 | 0.0416957 | -0.1340626 | 0.1049322 | 0.1866937 |
| 725253 | protein QIL1-like isoform 2 | 2.0037921 | -0.0001741 | 0.1135553 | -0.1522799 | 0.1897562 |
| GB50946 | uncharacterized protein LOC724626 | 0.7417706 | -0.1982565 | 0.1111897 | 0.2215719 | 0.1534713 |
| GB42823 | uncharacterized protein LOC100577440 isoform X3 | 0.7151644 | 0.1444905 | 0.3017972 | 0.0416428 | 0.2666875 |
Module 9
Table S34: List of all the genes in Module 9, ranked by their within-module connectivity, k. The latter four columns give the Log\(_2\) fold-change in expression in response to queen pheromone in each of the four species.
gene_list <- module_gene_list(9)
saveRDS(gene_list, file = "supplement/tab_S34.rds")
kable.table(gene_list)| Gene | Name | k | am_fc | bt_fc | lf_fc | ln_fc |
|---|---|---|---|---|---|---|
| GB49598 | RNA-binding protein Rsf1 | 3.4281703 | 0.3056885 | -0.0032899 | 0.1101417 | 0.0851146 |
| GB49355 | uncharacterized protein LOC100576266 isoform X2 | 3.2620545 | 0.3093869 | 0.0572808 | -0.1561239 | 0.1319324 |
| GB51008 | metaxin-2-like isoform 2 | 3.2002938 | 0.4439731 | -0.1469924 | 0.0424195 | 0.1446767 |
| GB52735 | DAZ-associated protein 2-like isoform X2 | 2.9721946 | 0.2871285 | 0.0231294 | 0.0623909 | -0.1752350 |
| GB55970 | proliferating cell nuclear antigen | 2.8130746 | 0.3776271 | 0.2351576 | 0.0776321 | 0.0776232 |
| 551833 | PAXIP1-associated glutamate-rich protein 1-like isoform 2 | 2.7565876 | 0.4962252 | 0.0926024 | 0.0590149 | 0.2492689 |
| GB43092 | cyclin-dependent kinase 5 | 2.5960809 | 0.3844390 | 0.0216430 | 0.0951021 | 0.1433308 |
| GB55381 | centrosomal protein of 97 kDa isoform X2 | 2.5582953 | 0.2955124 | -0.0548547 | 0.1869490 | 0.1357882 |
| GB43232 | transmembrane protein 222-like isoform 1 | 2.3398175 | 0.5861394 | 0.0458418 | 0.1933063 | 0.2422045 |
| GB50724 | peptidyl-tRNA hydrolase 2, mitochondrial-like isoform 1 | 2.2475846 | 0.7085344 | -0.0764084 | 0.0647428 | 0.1173188 |
| GB51226 | tyrosine-protein kinase CSK isoform X4 | 2.2239789 | 0.5181445 | 0.0036483 | 0.2021335 | 0.1570482 |
| GB56003 | methyltransferase-like protein 14 homolog | 2.2171705 | 0.3353470 | 0.1482768 | 0.1276493 | 0.1038143 |
| GB48128 | DNA-directed RNA polymerase III subunit RPC8-like isoform 1 | 2.1261420 | 0.7927715 | 0.1807240 | -0.1997985 | 0.1943152 |
| GB42319 | uncharacterized protein LOC409105 isoform 1 | 2.1081864 | 0.7613151 | -0.1786302 | 0.0944737 | 0.3063722 |
| GB45649 | adenosine 3’-phospho 5’-phosphosulfate transporter 1 | 2.0383947 | 0.5325883 | -0.0623392 | 0.1488054 | 0.2109308 |
| GB43086 | uncharacterized protein LOC726486 | 1.9692909 | 0.6676944 | 0.0549481 | 0.1238370 | 0.1069750 |
| GB45657 | cdc42 homolog isoform X2 | 1.9443700 | 0.4128119 | 0.1648899 | 0.1727575 | 0.0882596 |
| GB53270 | UPF0428 protein CXorf56 homolog isoformX2 | 1.9439132 | -0.0053964 | 0.0902614 | 0.0955928 | 0.1557436 |
| GB48852 | heterogeneous nuclear ribonucleoprotein H-like isoform X1 | 1.9171405 | 0.3943774 | -0.0242347 | 0.1867469 | 0.1629403 |
| GB45560 | 2-aminoethanethiol dioxygenase-like isoform X2 | 1.8928212 | 0.5646965 | 0.0858905 | 0.2044355 | 0.1964693 |
| GB53957 | U6 snRNA-associated Sm-like protein LSm1-like | 1.8732559 | 1.2758936 | 0.1485275 | 0.0654838 | 0.1025379 |
| GB55241 | myosin-9-like isoform X2 | 1.8577528 | 0.3826838 | 0.0273981 | 0.1613124 | 0.3355618 |
| GB42726 | lysosomal protein NCU-G1-A-like | 1.8538445 | 0.1961158 | -0.1161535 | 0.0827387 | 0.1842311 |
| GB52929 | soluble guanylyl cyclase alpha 1 subunit | 1.8202355 | 0.8632355 | 0.0679563 | 0.2472309 | 0.1991959 |
| GB55098 | progestin and adipoQ receptor family member 4-like isoform X3 | 1.8069386 | 1.0981696 | 0.0958375 | 0.2837369 | 0.3395738 |
| GB50885 | uncharacterized protein LOC409648 | 1.8009421 | 0.5217942 | -0.1408814 | 0.1076250 | -0.0919353 |
| GB54279 | cleavage and polyadenylation specificity factor subunit 4 | 1.7698409 | 0.3124242 | 0.3549593 | 1.6577375 | 0.2521921 |
| GB45810 | locomotion-related protein Hikaru genki isoform X4 | 1.7456427 | 0.4274473 | -0.0495554 | 0.1897961 | 0.1299238 |
| GB54147 | loss of heterozygosity 12 chromosomal region 1 protein homolog | 1.7319102 | 0.4424770 | -0.0518968 | 0.1119449 | 0.0806514 |
| GB48175 | probable cytochrome P450 305a1 | 1.7158930 | 1.5793489 | 0.0957169 | 0.0666737 | 0.2220252 |
| GB50090 | adenosine deaminase acting on RNA | 1.7017820 | 0.2845926 | -0.0201237 | 0.2675177 | 0.3162340 |
| GB55831 | aromatic-L-amino-acid decarboxylase isoform X2 | 1.6395161 | 0.7455077 | 0.7384633 | 0.0929703 | 0.2855078 |
| GB52236 | leucine-rich repeat-containing protein C10orf11 homolog isoform X1 | 1.6041255 | 0.2352825 | 0.0773345 | 0.0243112 | 0.1325469 |
| GB44143 | oxidative stress-induced growth inhibitor 1-like isoform X1 | 1.5781579 | 0.4074280 | 0.0477067 | 0.0904485 | 0.0514744 |
| GB42224 | leucine-rich repeat and calponin homology domain-containing protein 1-like isoform X2 | 1.5751039 | 0.4417497 | -0.0693594 | 1.2318066 | 0.2994614 |
| GB46734 | mitochondrial import inner membrane translocase subunit TIM14-like isoform X3 | 1.5718335 | 0.1173025 | 0.0125511 | 0.2021870 | 0.2728669 |
| GB43817 | atrial natriuretic peptide receptor 1-like | 1.5134486 | 0.4326071 | 0.1235643 | 0.1802794 | 0.0892441 |
| GB50722 | phospholipase A1 member A-like | 0.7527136 | 1.8995474 | 0.3289060 | -0.0744550 | 0.5503768 |
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] parallel stats4 grid stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] AnnotationHub_2.12.0 GO.db_3.6.0 AnnotationDbi_1.42.1
[4] IRanges_2.14.10 S4Vectors_0.18.3 Biobase_2.40.0
[7] BiocGenerics_0.26.0 fgsea_1.6.0 Rcpp_1.0.0
[10] clusterProfiler_3.8.1 kableExtra_0.9.0 pander_0.6.2
[13] sva_3.28.0 BiocParallel_1.14.2 genefilter_1.62.0
[16] mgcv_1.8-24 nlme_3.1-137 MuMIn_1.42.1
[19] ecodist_2.0.1 gplots_3.0.1 ggjoy_0.4.1
[22] ggridges_0.5.0 RColorBrewer_1.1-2 gridExtra_2.3
[25] ggdendro_0.1-20 ggrepel_0.8.0 ggplot2_3.1.0
[28] stringr_1.3.1 purrr_0.3.1 tidyr_0.8.2
[31] dplyr_0.8.0.1 reshape2_1.4.3 RSQLite_2.1.1
[34] WGCNA_1.63 fastcluster_1.1.25 dynamicTreeCut_1.63-1
loaded via a namespace (and not attached):
[1] backports_1.1.2 Hmisc_4.1-1
[3] fastmatch_1.1-0 workflowr_1.2.0
[5] plyr_1.8.4 igraph_1.2.1
[7] lazyeval_0.2.1 splines_3.5.1
[9] robust_0.4-18 digest_0.6.18
[11] BiocInstaller_1.30.0 foreach_1.4.4
[13] htmltools_0.3.6 GOSemSim_2.6.0
[15] viridis_0.5.1 gdata_2.18.0
[17] magrittr_1.5 checkmate_1.8.5
[19] memoise_1.1.0 fit.models_0.5-14
[21] cluster_2.0.7-1 doParallel_1.0.14
[23] limma_3.36.2 readr_1.1.1
[25] annotate_1.58.0 matrixStats_0.54.0
[27] enrichplot_1.0.2 colorspace_1.3-2
[29] blob_1.1.1 rvest_0.3.2
[31] rrcov_1.4-4 crayon_1.3.4
[33] RCurl_1.95-4.11 impute_1.54.0
[35] survival_2.42-6 iterators_1.0.10
[37] glue_1.3.0.9000 gtable_0.2.0
[39] UpSetR_1.3.3 DEoptimR_1.0-8
[41] scales_1.0.0 DOSE_3.6.1
[43] mvtnorm_1.0-8 DBI_1.0.0
[45] viridisLite_0.3.0 xtable_1.8-3
[47] htmlTable_1.12 units_0.6-0
[49] foreign_0.8-71 bit_1.1-14
[51] preprocessCore_1.42.0 Formula_1.2-3
[53] htmlwidgets_1.2 httr_1.3.1
[55] acepack_1.4.1 pkgconfig_2.0.2
[57] XML_3.98-1.12 nnet_7.3-12
[59] dbplyr_1.2.2 later_0.7.5
[61] tidyselect_0.2.5 labeling_0.3
[63] rlang_0.3.1 munsell_0.5.0
[65] tools_3.5.1 evaluate_0.11
[67] yaml_2.2.0 knitr_1.20
[69] bit64_0.9-7 fs_1.2.6
[71] robustbase_0.93-1.1 caTools_1.17.1.1
[73] ggraph_1.0.2 mime_0.6
[75] whisker_0.3-2 DO.db_2.9
[77] xml2_1.2.0 compiler_3.5.1
[79] rstudioapi_0.9.0 curl_3.2
[81] interactiveDisplayBase_1.18.0 tibble_2.0.99.9000
[83] tweenr_0.1.5 pcaPP_1.9-73
[85] stringi_1.3.1 highr_0.7
[87] lattice_0.20-35 Matrix_1.2-14
[89] pillar_1.3.1.9000 data.table_1.11.8
[91] cowplot_0.9.3 bitops_1.0-6
[93] httpuv_1.4.5 qvalue_2.12.0
[95] R6_2.4.0 latticeExtra_0.6-28
[97] promises_1.0.1 KernSmooth_2.23-15
[99] codetools_0.2-15 MASS_7.3-50
[101] gtools_3.8.1 assertthat_0.2.0
[103] rprojroot_1.3-2 withr_2.1.2
[105] hms_0.4.2 rpart_4.1-13
[107] rmarkdown_1.10 rvcheck_0.1.0
[109] git2r_0.23.0 ggforce_0.1.3
[111] shiny_1.2.0 base64enc_0.1-3